|
|
|
|
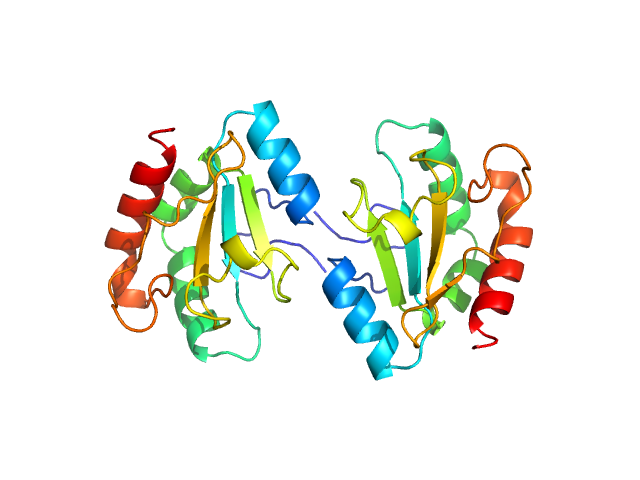
|
| Sample: |
Anterior gradient protein 2 homolog dimer, 30 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH7.4, 150 mM NaCl, 1% glycerol, 0.2 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2024 May 5
|
A structural basis for chaperone repression of stress signaling from the endoplasmic reticulum.
Mol Cell (2025)
Neidhardt L, Tung J, Kuchersky M, Milczarek J, Kargas V, Stott K, Rosenzweig R, Ron D, Yan Y
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.3 |
nm |
| VolumePorod |
30 |
nm3 |
|
|
|
|
|
|
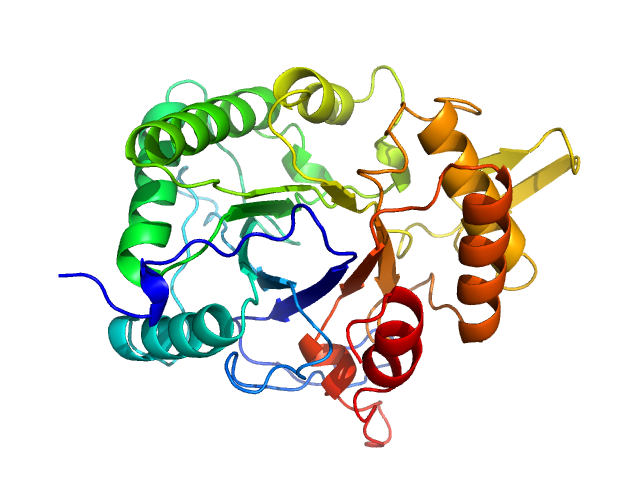
|
| Sample: |
Chitinase monomer, 36 kDa Listeria monocytogenes serovar … protein
|
| Buffer: |
20 mM Tris, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2024 Aug 4
|
Structure and analysis of a virulent chitinase from Listeria monocytogenes.
Biochim Biophys Acta Proteins Proteom :141106 (2025)
Rehman S, Baczynska M, Zhang B, Lorenz CD, Garnett JA
|
| RgGuinier |
2.2 |
nm |
| Dmax |
5.6 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
S9 peptidase form Rhizobium radiobacter monomer, 74 kDa Agrobacterium radiobacter protein
|
| Buffer: |
10 mM Tris-HCl, 135 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2024 Mar 21
|
Structural and functional characterization of S9 peptidase form Rhizobium radiobacter
Dr. Rahul Singh
|
| RgGuinier |
3.5 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
137 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
S9 peptidase form Rhizobium radiobacter monomer, 74 kDa Agrobacterium radiobacter protein
|
| Buffer: |
10 mM Tris-HCl, 135 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2024 Mar 21
|
Structural and functional characterization of S9 peptidase form Rhizobium radiobacter
Dr. Rahul Singh
|
| RgGuinier |
3.6 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
128 |
nm3 |
|
|
|
|
|
|
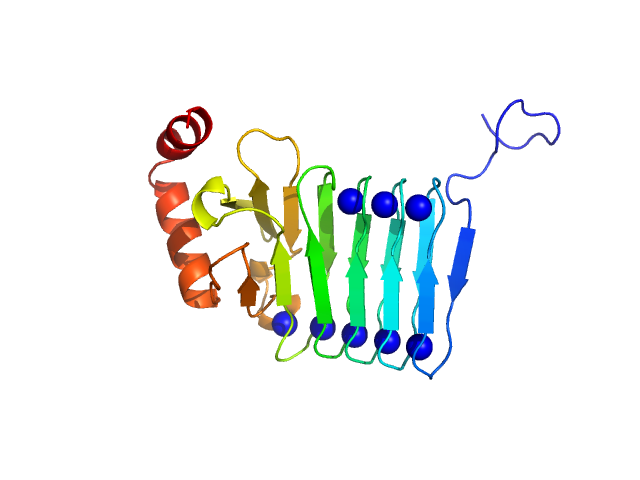
|
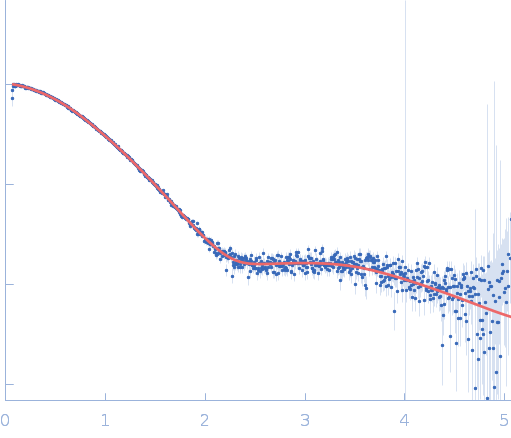
| Sample: |
Repeats-in-toxin domain Block V of adenylate cyclase toxin monomer, 19 kDa Bordetella pertussis protein
|
| Buffer: |
50 mM Tris, 5 mM DTT, 10 mM SrCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2024 Aug 1
|
Ion-selective conformational stabilization of a disordered repeats-in-toxin protein domain.
Biophys J (2025)
Gudinas AP, Shambharkar GM, Chang MP, Fernández D, Matsui T, Mai DJ
|
| RgGuinier |
2.1 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
36 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Repeats-in-toxin domain Block V of adenylate cyclase toxin monomer, 19 kDa Bordetella pertussis protein
|
| Buffer: |
50 mM Tris, 5 mM DTT , 3 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2024 Dec 12
|
Ion-selective conformational stabilization of a disordered repeats-in-toxin protein domain.
Biophys J (2025)
Gudinas AP, Shambharkar GM, Chang MP, Fernández D, Matsui T, Mai DJ
|
| RgGuinier |
2.0 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
|
|
|
|
|
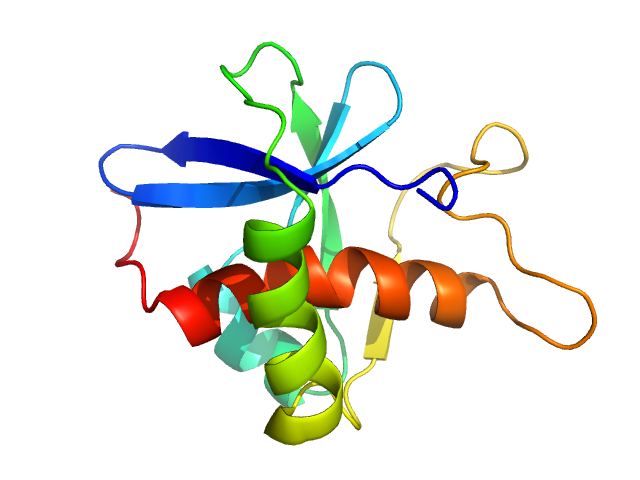
| Sample: |
CrTPpart2 of N-acetyl-gamma-glutamyl-phosphate reductase (ArgC) monomer, 14 kDa Paulinella chromatophora protein
|
| Buffer: |
20 mM HEPES, 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Sep 14
|
The Paulinella
chromatophore transit peptide part2 adopts a structural fold similar to the γ-glutamyl-cyclotransferase fold
Plant Physiology (2025)
Klimenko V, Reiners J, Applegate V, Reimann K, Popowicz G, Hoeppner A, Papadopoulos A, Smits S, Nowack E
|
| RgGuinier |
1.7 |
nm |
| Dmax |
5.8 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
|
|
|
|
|
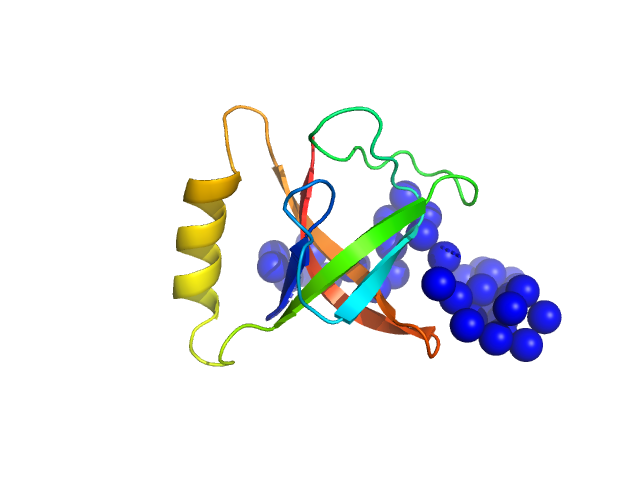
| Sample: |
Protein O-GlcNAcase dimer, 163 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris buffer, 150 mM NaCl, and 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at Bruker Nanostar w Excillum source, Department of Chemistry, iNANO building, Aarhus Uinversity on 2023 Jun 22
|
Multi-domain O-GlcNAcase structures reveal allosteric regulatory mechanisms
Nature Communications 16(1) (2025)
Hansen S, Bartual S, Yuan H, Raimi O, Gorelik A, Ferenbach A, Lytje K, Pedersen J, Drace T, Boesen T, van Aalten D
|
| RgGuinier |
5.4 |
nm |
| Dmax |
18.9 |
nm |
| VolumePorod |
443 |
nm3 |
|
|
|
|
|
|
|
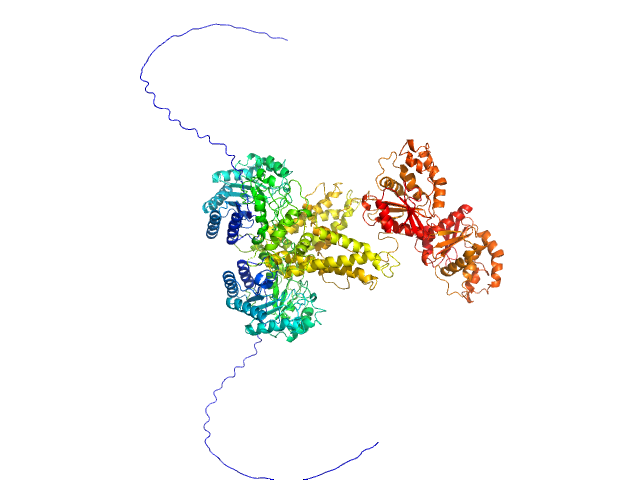
| Sample: |
Genome polyprotein monomer, 13 kDa Turkey avisivirus protein
|
| Buffer: |
20 mM citrate pH 5.4, 150 mM NaCl, 2 mM TCEP, 1% glycerol, pH: 5.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 May 2
|
Solution structure and functional characterization of an avian picornavirus 2AH/NC protein
Anna Wehlin
|
| RgGuinier |
1.7 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|
|
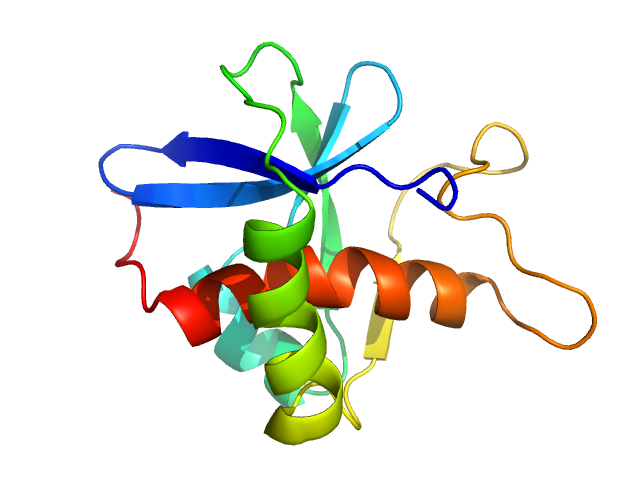
| Sample: |
Genome polyprotein monomer, 13 kDa Turkey avisivirus protein
|
| Buffer: |
20mM Citrate buffer pH 6.5, 150mM NaCl, 2mM TCEP, 1% glycerol, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 May 15
|
Solution structure and functional characterization of an avian picornavirus 2AH/NC protein
Anna Wehlin
|
| RgGuinier |
1.6 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
27 |
nm3 |
|
|