|
|
|
|
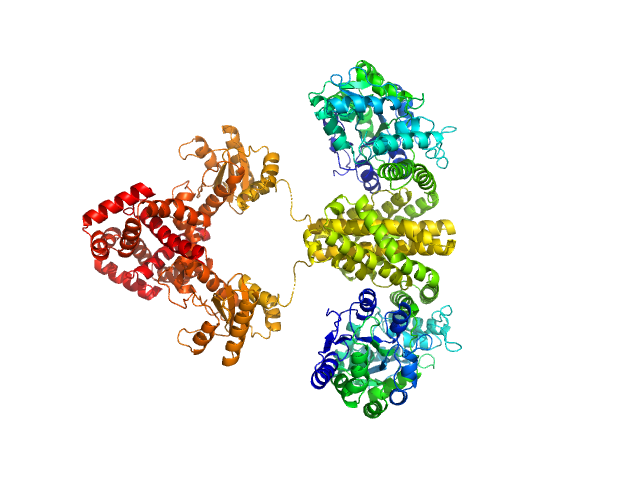
|
| Sample: |
Maltose/maltodextrin-binding periplasmic protein (D108A, K109A, E198A, N199A, K265A) dimer, 80 kDa Escherichia coli (strain … protein
Uncharacterized protein Rv2242 dimer, 86 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50 mM Tris pH 7.5, 150 mM NaCl, pH: |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2024 Apr 3
|
Domain architecture of the Mycobacterium tuberculosis MabR (Rv2242), a member of the PucR transcription factor family
Heliyon :e40494 (2024)
Megalizzi V, Tanina A, Grosse C, Mirgaux M, Legrand P, Mirandela G, Wohlkönig A, Bifani P, Wintjens R
|
| RgGuinier |
3.7 |
nm |
| Dmax |
10.8 |
nm |
| VolumePorod |
345 |
nm3 |
|
|
|
|
|
|
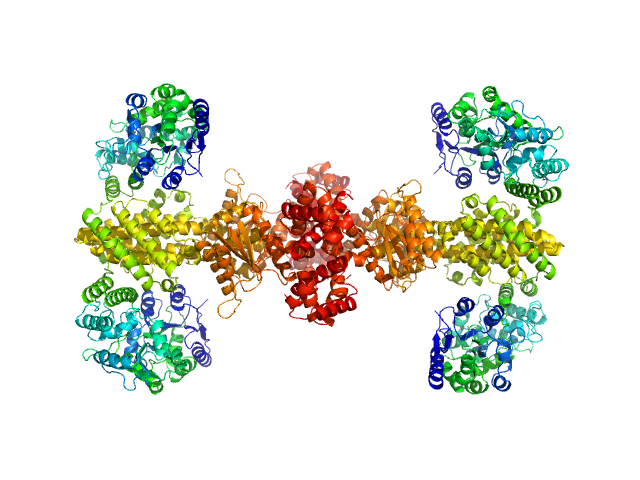
|
| Sample: |
Maltose/maltodextrin-binding periplasmic protein (D108A, K109A, E198A, N199A, K265A) tetramer, 161 kDa Escherichia coli (strain … protein
Uncharacterized protein Rv2242 tetramer, 171 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50 mM Tris pH 7.5, 150 mM NaCl, pH: |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2024 Apr 3
|
Domain architecture of the Mycobacterium tuberculosis MabR (Rv2242), a member of the PucR transcription factor family
Heliyon :e40494 (2024)
Megalizzi V, Tanina A, Grosse C, Mirgaux M, Legrand P, Mirandela G, Wohlkönig A, Bifani P, Wintjens R
|
| RgGuinier |
5.2 |
nm |
| Dmax |
15.8 |
nm |
| VolumePorod |
826 |
nm3 |
|
|
|
|
|
|
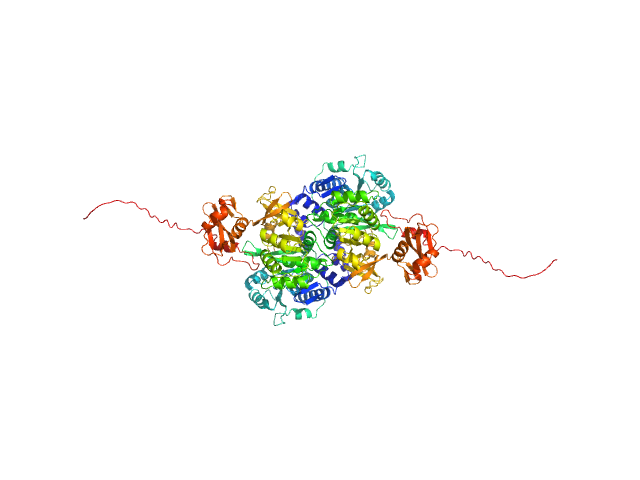
|
| Sample: |
Probable fatty-acid-CoA ligase FadD5 (Fatty-acid-CoA synthetase) (Fatty-acid-CoA synthase) dimer, 123 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM HEPES, 500 mM NaCl, 5 mM MgCl2, 1 mM β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Oct 20
|
Structural enzymological studies of the long chain fatty acyl-CoA synthetase FadD5 from the mce1 operon of Mycobacterium tuberculosis.
Biochem Biophys Res Commun 769:151960 (2025)
Rahman MA, Dalwani S, Venkatesan R
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.3 |
nm |
| VolumePorod |
221 |
nm3 |
|
|
|
|
|
|
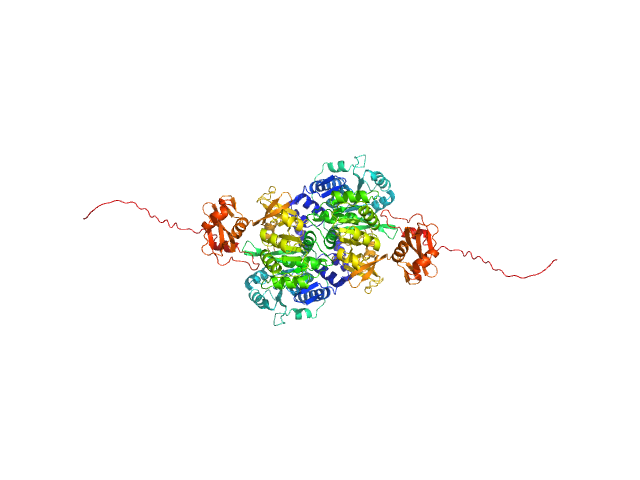
|
| Sample: |
Probable fatty-acid-CoA ligase FadD5 (Fatty-acid-CoA synthetase) (Fatty-acid-CoA synthase) dimer, 123 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM HEPES, 500 mM NaCl, 5 mM MgCl2, 1 mM β-mercaptoethanol, 2mM ATP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Oct 20
|
Structural enzymological studies of the long chain fatty acyl-CoA synthetase FadD5 from the mce1 operon of Mycobacterium tuberculosis.
Biochem Biophys Res Commun 769:151960 (2025)
Rahman MA, Dalwani S, Venkatesan R
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
213 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Probable fatty-acid-CoA ligase FadD5 (Fatty-acid-CoA synthetase) (Fatty-acid-CoA synthase) dimer, 123 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM HEPES, 500 mM NaCl, 5 mM MgCl2, 1 mM β-mercaptoethanol, 2 mM coenzyme A (CoA), pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Oct 20
|
Structural enzymological studies of the long chain fatty acyl-CoA synthetase FadD5 from the mce1 operon of Mycobacterium tuberculosis.
Biochem Biophys Res Commun 769:151960 (2025)
Rahman MA, Dalwani S, Venkatesan R
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
210 |
nm3 |
|
|
|
|
|
|
|
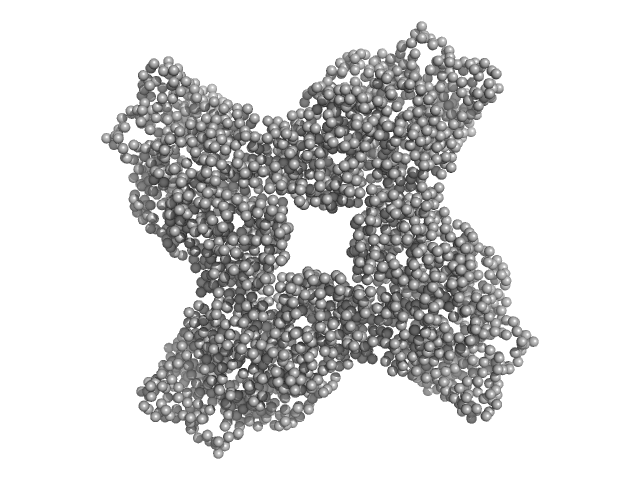
| Sample: |
Putative acylaminoacyl-peptidase tetramer, 293 kDa Bacillus spizizenii (strain … protein
|
| Buffer: |
10 mM Tris-HCl, 135 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2024 Mar 18
|
Structural adaptations for carboxypeptidase activity in putative S9 acylaminoacyl peptidase from Bacillus subtilis.
Int J Biol Macromol :136734 (2024)
Chandravanshi K, Singh R, Kumar A, Bhange GN, Kumar A, Makde RD
|
| RgGuinier |
5.2 |
nm |
| Dmax |
15.9 |
nm |
| VolumePorod |
439 |
nm3 |
|
|
|
|
|
|
|
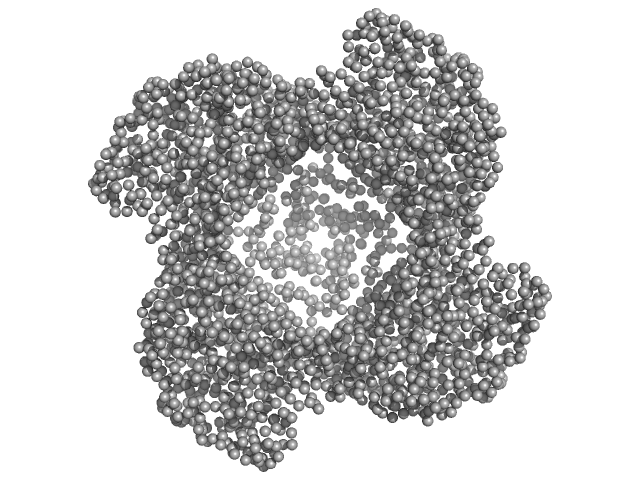
| Sample: |
Putative acylaminoacyl-peptidase tetramer, 293 kDa Bacillus spizizenii (strain … protein
|
| Buffer: |
10 mM Tris-HCl, 135 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2024 Mar 18
|
Structural adaptations for carboxypeptidase activity in putative S9 acylaminoacyl peptidase from Bacillus subtilis.
Int J Biol Macromol :136734 (2024)
Chandravanshi K, Singh R, Kumar A, Bhange GN, Kumar A, Makde RD
|
| RgGuinier |
5.2 |
nm |
| Dmax |
14.7 |
nm |
| VolumePorod |
435 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Putative acylaminoacyl-peptidase tetramer, 293 kDa Bacillus spizizenii (strain … protein
|
| Buffer: |
10 mM Tris-HCl, 135 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2024 Mar 18
|
Structural adaptations for carboxypeptidase activity in putative S9 acylaminoacyl peptidase from Bacillus subtilis.
Int J Biol Macromol :136734 (2024)
Chandravanshi K, Singh R, Kumar A, Bhange GN, Kumar A, Makde RD
|
| RgGuinier |
5.2 |
nm |
| Dmax |
15.3 |
nm |
| VolumePorod |
446 |
nm3 |
|
|
|
|
|
|
|
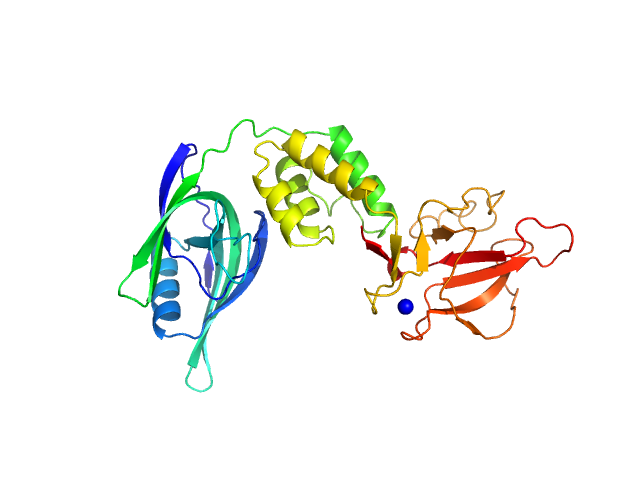
| Sample: |
Cereblon-midi monomer, 37 kDa protein
|
| Buffer: |
20 mM Hepes, 500 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Mar 1
|
Design of a Cereblon construct for crystallographic and biophysical studies of protein degraders
Nature Communications 15(1) (2024)
Kroupova A, Spiteri V, Rutter Z, Furihata H, Darren D, Ramachandran S, Chakraborti S, Haubrich K, Pethe J, Gonzales D, Wijaya A, Rodriguez-Rios M, Sturbaut M, Lynch D, Farnaby W, Nakasone M, Zollman D, Ciulli A
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.6 |
nm |
| VolumePorod |
61 |
nm3 |
|
|
|
|
|
|
|
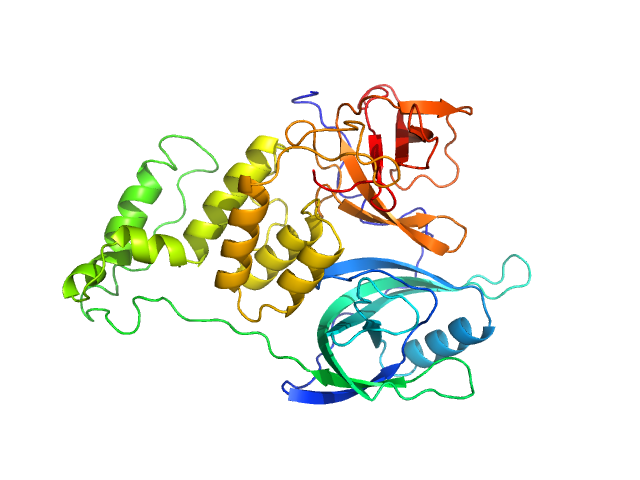
| Sample: |
Cereblon-midi monomer, 37 kDa protein
Mezigdomide monomer, 1 kDa synthetic construct
|
| Buffer: |
20 mM Hepes, 500 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Mar 1
|
Design of a Cereblon construct for crystallographic and biophysical studies of protein degraders
Nature Communications 15(1) (2024)
Kroupova A, Spiteri V, Rutter Z, Furihata H, Darren D, Ramachandran S, Chakraborti S, Haubrich K, Pethe J, Gonzales D, Wijaya A, Rodriguez-Rios M, Sturbaut M, Lynch D, Farnaby W, Nakasone M, Zollman D, Ciulli A
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
60 |
nm3 |
|
|