|
|
|
|

|
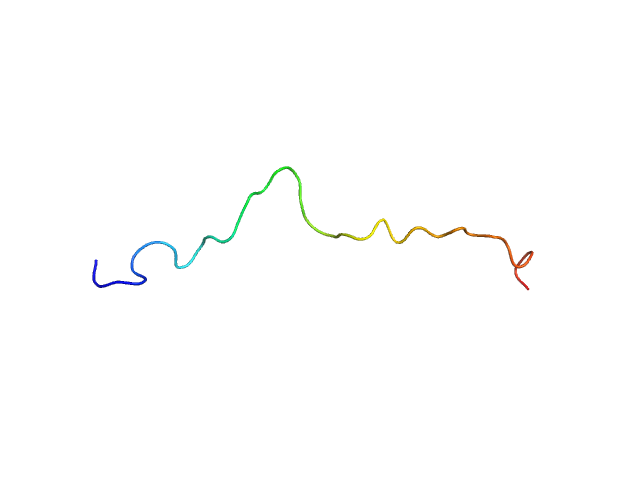
| Sample: |
NT17 monomer, 2 kDa synthetic construct protein
|
| Buffer: |
Sodium phosphate buffer, pH: 7.4 |
| Experiment: |
SAXS
data collected at TPS13A, NSRRC on 2021 Oct 21
|
Binding structures of SERF1a with NT17-polyQ peptides of huntingtin exon 1 revealed by SEC-SWAXS, NMR and molecular simulation.
IUCrJ (2024)
Lin TC, Shih O, Tsai TY, Yeh YQ, Liao KF, Mansel BW, Shiu YJ, Chang CF, Su AC, Chen YR, Jeng US
|
| RgGuinier |
1.2 |
nm |
| Dmax |
4.0 |
nm |
| VolumePorod |
5 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
HTT0 monomer, 4 kDa synthetic construct protein
|
| Buffer: |
Sodium phosphate buffer, pH: 7.4 |
| Experiment: |
SAXS
data collected at TPS13A, NSRRC on 2023 Nov 10
|
Binding structures of SERF1a with NT17-polyQ peptides of huntingtin exon 1 revealed by SEC-SWAXS, NMR and molecular simulation.
IUCrJ (2024)
Lin TC, Shih O, Tsai TY, Yeh YQ, Liao KF, Mansel BW, Shiu YJ, Chang CF, Su AC, Chen YR, Jeng US
|
| RgGuinier |
1.5 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
4 |
nm3 |
|
|
|
|
|
|
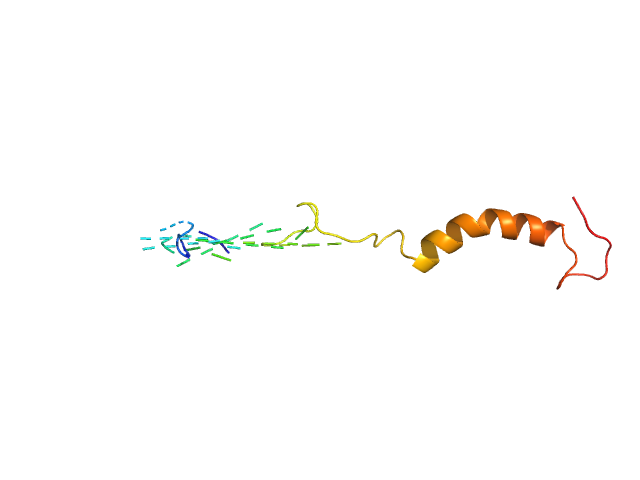
|
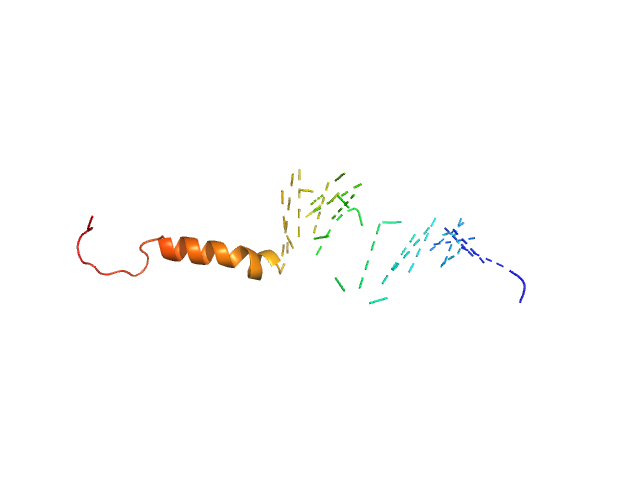
| Sample: |
Isoform Short of Small EDRK-rich factor 1 monomer, 7 kDa Homo sapiens protein
NT17 dimer, 4 kDa synthetic construct protein
|
| Buffer: |
Sodium phosphate buffer, pH: 7.4 |
| Experiment: |
SAXS
data collected at TPS13A, NSRRC on 2021 Oct 21
|
Binding structures of SERF1a with NT17-polyQ peptides of huntingtin exon 1 revealed by SEC-SWAXS, NMR and molecular simulation.
IUCrJ (2024)
Lin TC, Shih O, Tsai TY, Yeh YQ, Liao KF, Mansel BW, Shiu YJ, Chang CF, Su AC, Chen YR, Jeng US
|
|
|
|
|
|
|
|
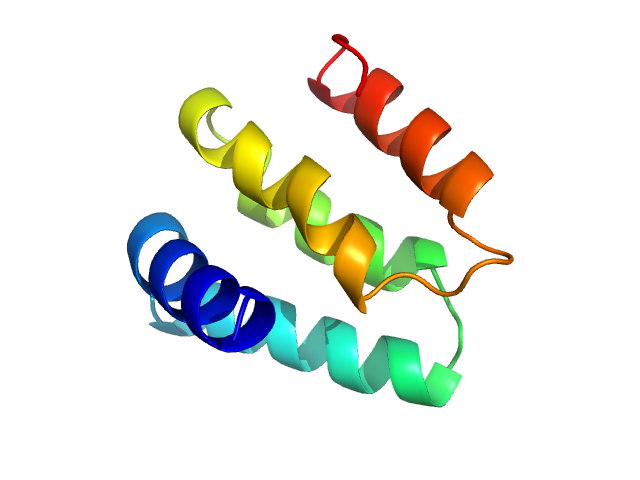
| Sample: |
Isoform Short of Small EDRK-rich factor 1 monomer, 7 kDa Homo sapiens protein
HTT3 monomer, 4 kDa synthetic construct protein
|
| Buffer: |
Sodium phosphate buffer, pH: 7.4 |
| Experiment: |
SAXS
data collected at TPS13A, NSRRC on 2021 May 20
|
Binding structures of SERF1a with NT17-polyQ peptides of huntingtin exon 1 revealed by SEC-SWAXS, NMR and molecular simulation.
IUCrJ (2024)
Lin TC, Shih O, Tsai TY, Yeh YQ, Liao KF, Mansel BW, Shiu YJ, Chang CF, Su AC, Chen YR, Jeng US
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
9 |
nm3 |
|
|
|
|
|
|
|
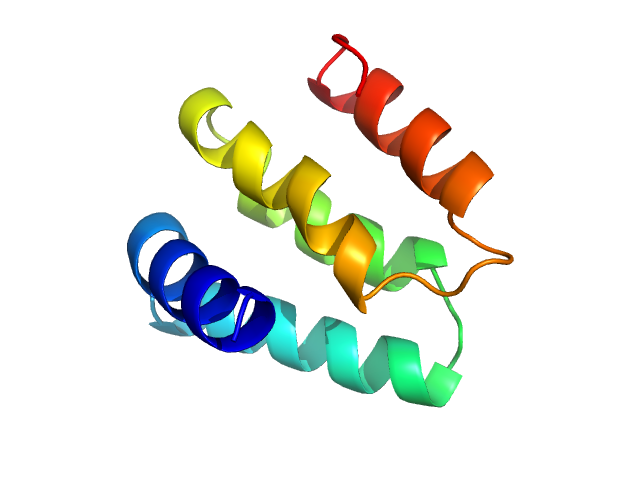
| Sample: |
Regulator of telomere elongation helicase 1 (Isoform 6, 1053-1147) monomer, 11 kDa Homo sapiens protein
|
| Buffer: |
50 mM Hepes, 150 mM NaCl,10 (v/v)% glycerol, 1 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Mar 22
|
Structural and biochemical characterization of the C‐terminal region of the human RTEL
1 helicase
Protein Science 33(9) (2024)
Cortone G, Graewert M, Kanade M, Longo A, Hegde R, González‐Magaña A, Chaves‐Arquero B, Blanco F, Napolitano L, Onesti S
|
| RgGuinier |
1.5 |
nm |
| Dmax |
5.0 |
nm |
| VolumePorod |
24 |
nm3 |
|
|
|
|
|
|
|
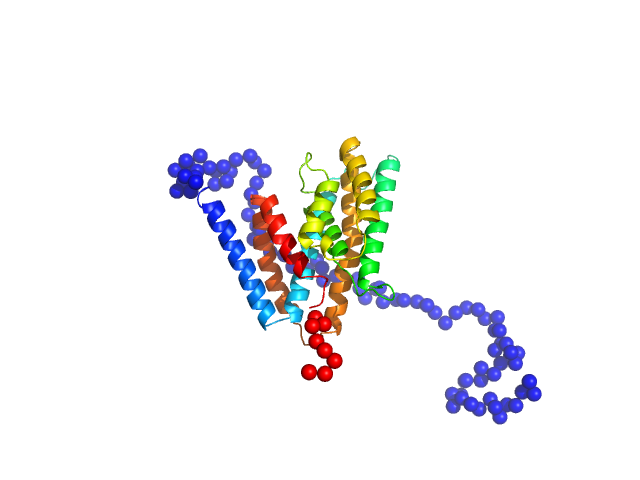
| Sample: |
Regulator of telomere elongation helicase 1 (Isoform 6, 889-974) monomer, 10 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl,10 (v/v)% glycerol, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Mar 22
|
Structural and biochemical characterization of the C‐terminal region of the human RTEL
1 helicase
Protein Science 33(9) (2024)
Cortone G, Graewert M, Kanade M, Longo A, Hegde R, González‐Magaña A, Chaves‐Arquero B, Blanco F, Napolitano L, Onesti S
|
| RgGuinier |
1.5 |
nm |
| Dmax |
4.6 |
nm |
| VolumePorod |
22 |
nm3 |
|
|
|
|
|
|
|
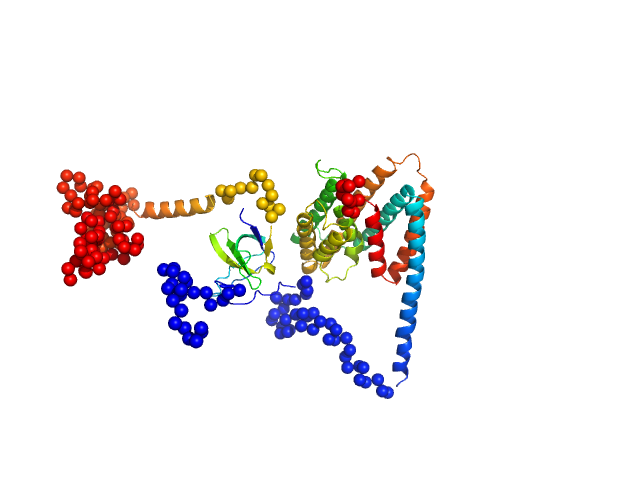
| Sample: |
Uncharacterized protein monomer, 35 kDa Chlamydia pneumoniae protein
|
| Buffer: |
137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, 3 %Glycerol, pH: 8.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Dec 14
|
The Chlamydia pneumoniae effector SemD exploits its host’s endocytic machinery by structural and functional mimicry
Nature Communications 15(1) (2024)
Kocher F, Applegate V, Reiners J, Port A, Spona D, Hänsch S, Mirzaiebadizi A, Ahmadian M, Smits S, Hegemann J, Mölleken K
|
| RgGuinier |
2.8 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
67 |
nm3 |
|
|
|
|
|
|
|
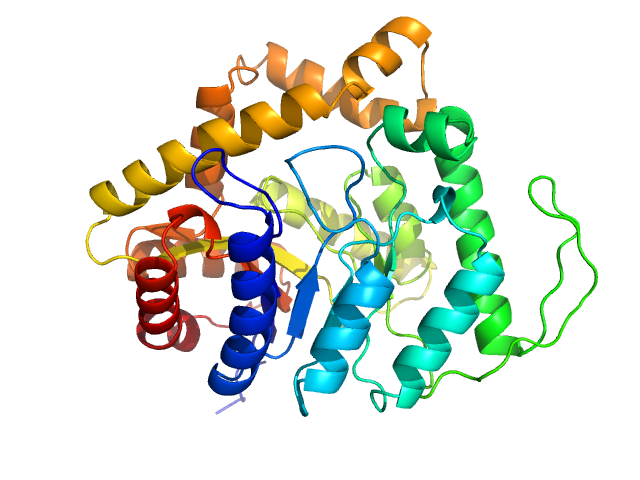
| Sample: |
Uncharacterized protein monomer, 35 kDa Chlamydia pneumoniae protein
Sorting nexin-9 monomer, 19 kDa Homo sapiens protein
|
| Buffer: |
137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, 3 %Glycerol, pH: 8.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Mar 20
|
The Chlamydia pneumoniae effector SemD exploits its host’s endocytic machinery by structural and functional mimicry
Nature Communications 15(1) (2024)
Kocher F, Applegate V, Reiners J, Port A, Spona D, Hänsch S, Mirzaiebadizi A, Ahmadian M, Smits S, Hegemann J, Mölleken K
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
94 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Alkanal monooxygenase alpha chain dimer, 84 kDa Enhygromyxa salina protein
|
| Buffer: |
150 mM NaCl, 25 mM Tris, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2020 Aug 1
|
luxA
Gene From Enhygromyxa salina
Encodes a Functional Homodimeric Luciferase
Proteins: Structure, Function, and Bioinformatics (2024)
Yudenko A, Bazhenov S, Aleksenko V, Goncharov I, Semenov O, Remeeva A, Nazarenko V, Kuznetsova E, Fomin V, Konopleva M, Al Ebrahim R, Sluchanko N, Ryzhykau Y, Semenov Y, Kuklin A, Manukhov I, Gushchin I
|
| RgGuinier |
3.3 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
138 |
nm3 |
|
|
|
|
|
|
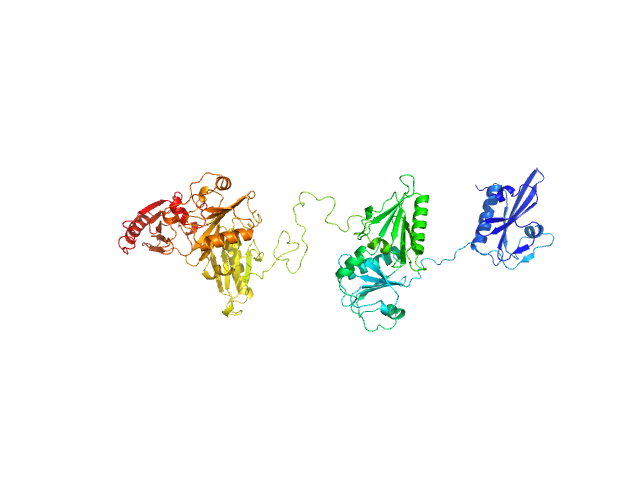
|
| Sample: |
Gelsolin monomer, 84 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris-HCl, pH8, 45 mM NaCl, 1 mM EGTA + 4 mM CaCl2, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Aug 31
|
Visualizing the nucleating and capped states of f-actin by Ca(2+)-gelsolin: Saxs data based structures of binary and ternary complexes.
Int J Biol Macromol :134556 (2024)
Sagar A, Peddada N, Choudhary V, Mir Y, Garg R, Ashish
|
| RgGuinier |
4.2 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
129 |
nm3 |
|
|