|
|
|
|
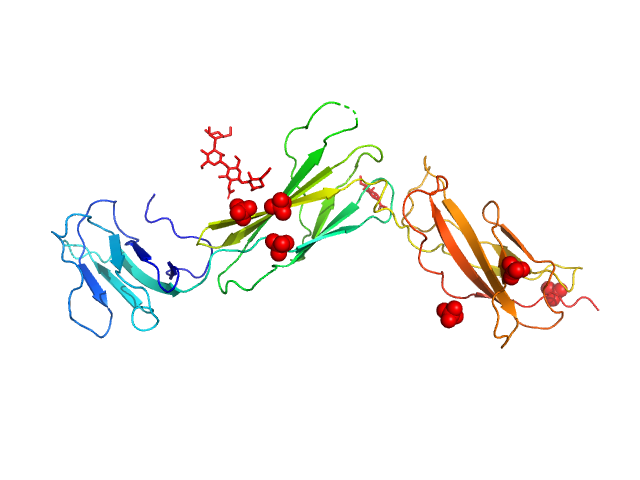
|
| Sample: |
Interleukin-11 receptor subunit alpha monomer, 32 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.2% sodium azide, pH: 8.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Jun 8
|
The structure of the extracellular domains of human interleukin 11 α-receptor reveals mechanisms of cytokine engagement
Journal of Biological Chemistry :jbc.RA119.012351 (2020)
Metcalfe R, Aizel K, Zlatic C, Nguyen P, Morton C, Lio D, Cheng H, Dobson R, Parker M, Gooley P, Putoczki T, Griffin M
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|
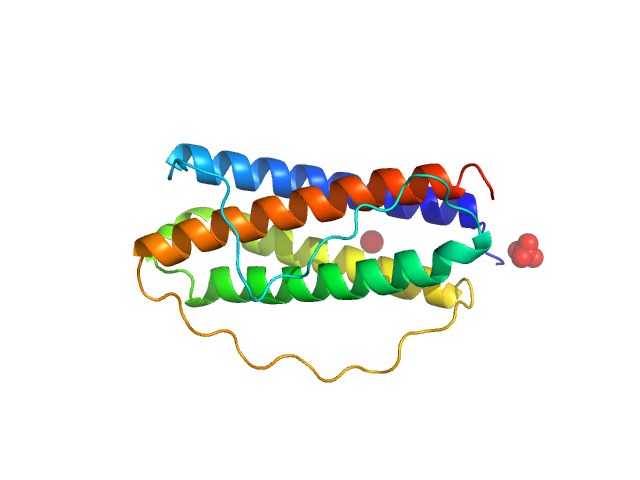
|
| Sample: |
Interleukin 11 monomer, 18 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.2% sodium azide, pH: 8.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Jun 8
|
The structure of the extracellular domains of human interleukin 11 α-receptor reveals mechanisms of cytokine engagement
Journal of Biological Chemistry :jbc.RA119.012351 (2020)
Metcalfe R, Aizel K, Zlatic C, Nguyen P, Morton C, Lio D, Cheng H, Dobson R, Parker M, Gooley P, Putoczki T, Griffin M
|
| RgGuinier |
1.7 |
nm |
| Dmax |
5.4 |
nm |
| VolumePorod |
22 |
nm3 |
|
|
|
|
|
|
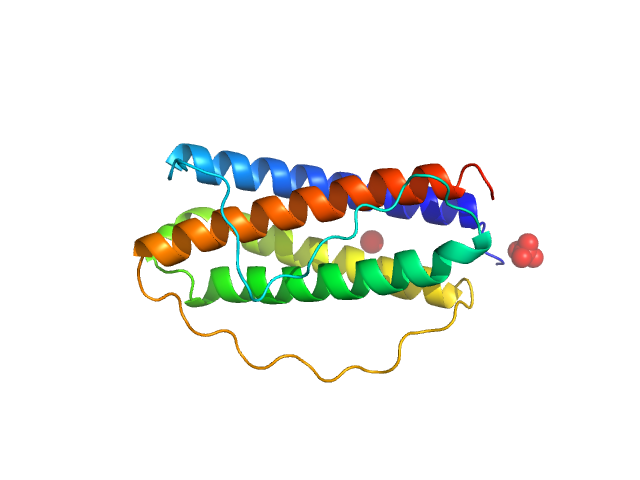
|
| Sample: |
Interleukin 11 monomer, 19 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.2% sodium azide, pH: 8.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Jun 8
|
The structure of the extracellular domains of human interleukin 11 α-receptor reveals mechanisms of cytokine engagement
Journal of Biological Chemistry :jbc.RA119.012351 (2020)
Metcalfe R, Aizel K, Zlatic C, Nguyen P, Morton C, Lio D, Cheng H, Dobson R, Parker M, Gooley P, Putoczki T, Griffin M
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.1 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|
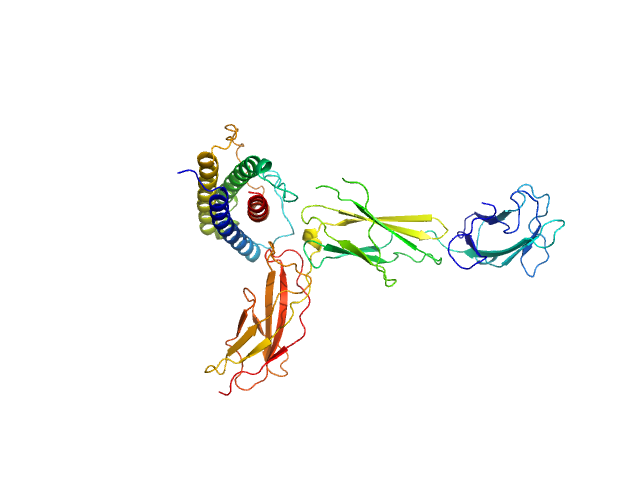
|
| Sample: |
Interleukin-11 receptor subunit alpha monomer, 32 kDa Homo sapiens protein
Interleukin 11 monomer, 18 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.2% sodium azide, pH: 8.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Jun 8
|
The structure of the extracellular domains of human interleukin 11 α-receptor reveals mechanisms of cytokine engagement
Journal of Biological Chemistry :jbc.RA119.012351 (2020)
Metcalfe R, Aizel K, Zlatic C, Nguyen P, Morton C, Lio D, Cheng H, Dobson R, Parker M, Gooley P, Putoczki T, Griffin M
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.2 |
nm |
| VolumePorod |
84 |
nm3 |
|
|
|
|
|
|
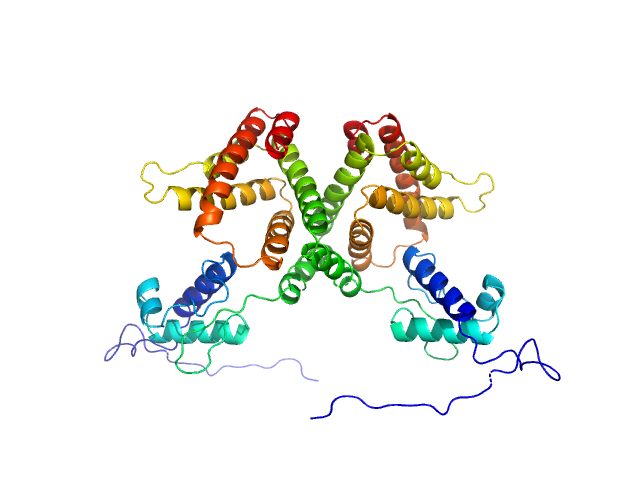
|
| Sample: |
HTH-type transcriptional repressor NanR dimer, 59 kDa Escherichia coli protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.1 % (w/v) sodium azide, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Apr 20
|
Mechanism of NanR gene repression and allosteric induction of bacterial sialic acid metabolism
(2020)
Horne C, Venugopal H, Panjikar S, Henrickson A, Brookes E, North R, Murphy J, Friemann R, Griffin M, Ramm G, Demeler B, Dobson R
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
110 |
nm3 |
|
|
|
|
|
|
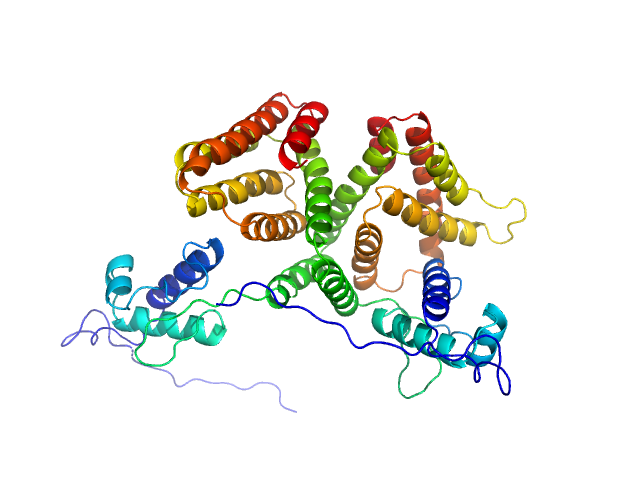
|
| Sample: |
HTH-type transcriptional repressor NanR dimer, 59 kDa Escherichia coli protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 20 mM Neu5Ac and 0.1 % (w/v) sodium azide, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Apr 20
|
Mechanism of NanR gene repression and allosteric induction of bacterial sialic acid metabolism
(2020)
Horne C, Venugopal H, Panjikar S, Henrickson A, Brookes E, North R, Murphy J, Friemann R, Griffin M, Ramm G, Demeler B, Dobson R
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
105 |
nm3 |
|
|
|
|
|
|
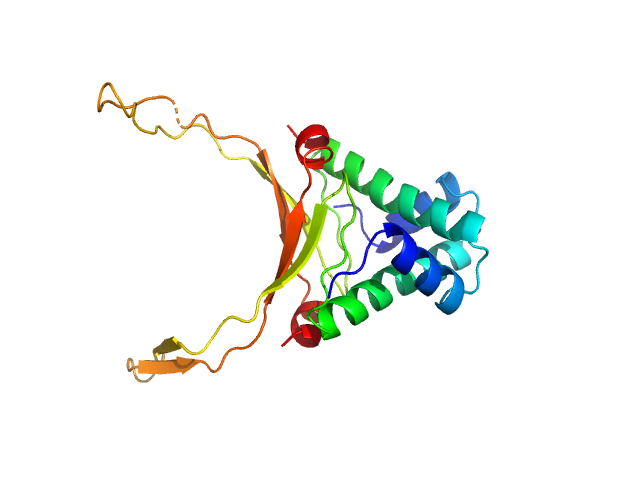
|
| Sample: |
DNA-binding protein HU-alpha, E38K/V42L double mutant decamer, 95 kDa Escherichia coli protein
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, 1 mM DTT, 1 mM PMSF, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2015 Apr 23
|
Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling (supplementary)
Soumya G Remesh
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
53 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Phosphoprotein tetramer, 317 kDa Nipah henipavirus protein
|
| Buffer: |
20 mM Tris-HCL, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at ID14-3, ESRF on 2011 Nov 3
|
Structural Description of the Nipah Virus Phosphoprotein and Its Interaction with STAT1.
Biophys J (2020)
Jensen MR, Yabukarski F, Communie G, Condamine E, Mas C, Volchkova V, Tarbouriech N, Bourhis JM, Volchkov V, Blackledge M, Jamin M
|
| RgGuinier |
10.9 |
nm |
| Dmax |
39.3 |
nm |
|
|
|
|
|
|

|
| Sample: |
Ile-Leu-Gln-Ile-Asn-Ser peptide, 1 kDa synthetic construct protein
|
| Buffer: |
pure (MQ, 18 MΩ) Water, pH: 7 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Nov 1
|
Amyloid Evolution: Antiparallel Replaced by Parallel.
Biophys J (2020)
Zanjani AAH, Reynolds NP, Zhang A, Schilling T, Mezzenga R, Berryman JT
|
|
|
|
|
|
|
|
| Sample: |
Ile-Leu-Gln-Ile-Asn-Ser peptide, 1 kDa synthetic construct protein
|
| Buffer: |
pure (MQ, 18 MΩ) Water, pH: 7 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Nov 1
|
Amyloid Evolution: Antiparallel Replaced by Parallel.
Biophys J (2020)
Zanjani AAH, Reynolds NP, Zhang A, Schilling T, Mezzenga R, Berryman JT
|
|
|