|
|
|
|
|
| Sample: |
Probable exodeoxyribonuclease III protein XthA monomer, 33 kDa Mycobacterium tuberculosis protein
M. tb. LigA BRCT domain monomer, 15 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
50 mM Tris-HCl 500 mM NaCl 5mM β-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Mar 9
|
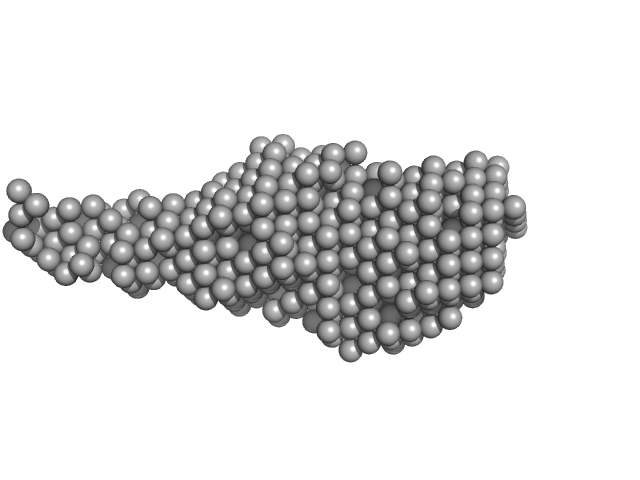
M. tuberculosis class II apurinic/ apyrimidinic-endonuclease/3'-5' exonuclease (XthA) engages with NAD+-dependent DNA ligase A (LigA) to counter futile cleavage and ligation cycles in base excision repair.
Nucleic Acids Res (2020)
Khanam T, Afsar M, Shukla A, Alam F, Kumar S, Soyar H, Dolma K, Pasupuleti M, Srivastava KK, Ampapathi RS, Ramachandran R
|
| RgGuinier |
3.7 |
nm |
| Dmax |
18.5 |
nm |
| VolumePorod |
112 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
DNA ligase A monomer, 76 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
50 mM Tris-HCl, 200 mM NaCl, 2 mM β-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 May 13
|
M. tuberculosis class II apurinic/ apyrimidinic-endonuclease/3'-5' exonuclease (XthA) engages with NAD+-dependent DNA ligase A (LigA) to counter futile cleavage and ligation cycles in base excision repair.
Nucleic Acids Res (2020)
Khanam T, Afsar M, Shukla A, Alam F, Kumar S, Soyar H, Dolma K, Pasupuleti M, Srivastava KK, Ampapathi RS, Ramachandran R
|
| RgGuinier |
5.2 |
nm |
| Dmax |
16.7 |
nm |
| VolumePorod |
870 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA ligase A monomer, 76 kDa Mycobacterium tuberculosis protein
Probable exodeoxyribonuclease III protein XthA monomer, 33 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
50 mM Tris-HCl, 200 mM NaCl, 2 mM β-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 May 13
|
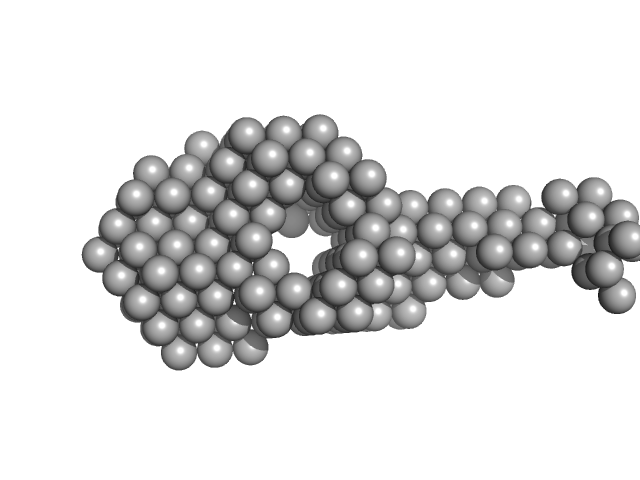
M. tuberculosis class II apurinic/ apyrimidinic-endonuclease/3'-5' exonuclease (XthA) engages with NAD+-dependent DNA ligase A (LigA) to counter futile cleavage and ligation cycles in base excision repair.
Nucleic Acids Res (2020)
Khanam T, Afsar M, Shukla A, Alam F, Kumar S, Soyar H, Dolma K, Pasupuleti M, Srivastava KK, Ampapathi RS, Ramachandran R
|
| RgGuinier |
6.2 |
nm |
| Dmax |
23.9 |
nm |
| VolumePorod |
662 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Somapacitan monomer, 22 kDa Homo sapiens protein
FcRn binding optimised human serum albumin V418M, T420A, E505G, V547A monomer, 66 kDa Homo sapiens protein
Neonatal Fc receptor ectodomain beta-microglogulin part with C-terminal His6 tag monomer, 13 kDa Homo sapiens protein
Neonatal Fc receptor ectodomain alpha-chain monomer, 30 kDa Homo sapiens protein
|
| Buffer: |
100 mM MES, 100 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at I911-4, MAX IV on 2015 May 11
|
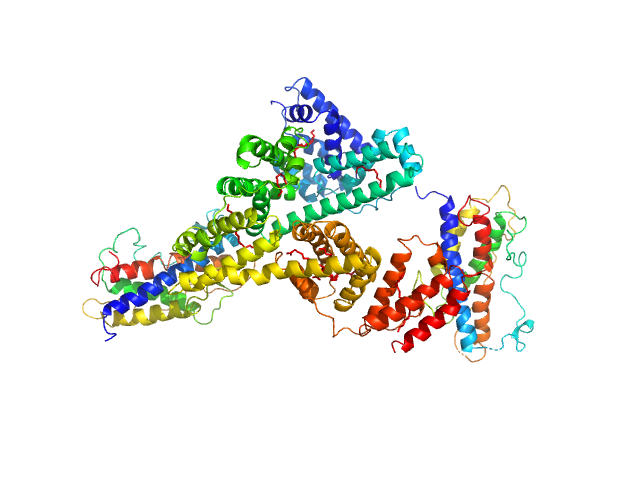
Identification of binding sites on human serum albumin for somapacitan - a long-acting growth hormone derivative.
Biochemistry (2020)
Johansson E, Nielsen AD, Demuth H, Wiberg C, Schjødt C, Huang T, Chen J, Jensen S, Petersen J, Thygesen P
|
| RgGuinier |
4.2 |
nm |
| Dmax |
14.7 |
nm |
| VolumePorod |
227 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
FcRn binding optimised human serum albumin V418M, T420A, E505G, V547A monomer, 66 kDa Homo sapiens protein
Neonatal Fc receptor ectodomain beta-microglogulin part with C-terminal His6 tag monomer, 13 kDa Homo sapiens protein
Albumin-binding side-chain monomer, 1 kDa
Neonatal Fc receptor ectodomain alpha-chain monomer, 30 kDa Homo sapiens protein
|
| Buffer: |
100 mM MES, 100 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at I911-4, MAX IV on 2015 Nov 11
|
Identification of binding sites on human serum albumin for somapacitan - a long-acting growth hormone derivative.
Biochemistry (2020)
Johansson E, Nielsen AD, Demuth H, Wiberg C, Schjødt C, Huang T, Chen J, Jensen S, Petersen J, Thygesen P
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.6 |
nm |
| VolumePorod |
174 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Human serum albumin monomer, 66 kDa Homo sapiens protein
Somapacitan dimer, 44 kDa Homo sapiens protein
|
| Buffer: |
100 mM MES, 140 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, Novo Nordisk A/S on 2015 Sep 4
|
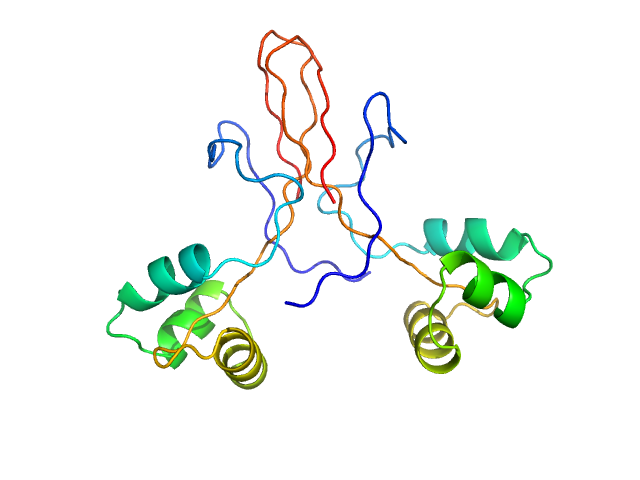
Identification of binding sites on human serum albumin for somapacitan - a long-acting growth hormone derivative.
Biochemistry (2020)
Johansson E, Nielsen AD, Demuth H, Wiberg C, Schjødt C, Huang T, Chen J, Jensen S, Petersen J, Thygesen P
|
| RgGuinier |
4.1 |
nm |
| Dmax |
13.9 |
nm |
| VolumePorod |
202 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Protein ninH dimer, 20 kDa Escherichia phage lambda protein
|
| Buffer: |
150 mM NaCl, 50 mM phosphate buffer (pH 7.4), 1 mM EDTA, pH: 7.4 |
| Experiment: |
SAXS
data collected at Xenocs Xeuss 2.0 with MetalJet, Department of Macromolecular Physics, Faculty of Physics, Adam Mickiewicz University on 2019 Jul 1
|
A bacteriophage mimic of the bacterial nucleoid-associated protein Fis.
Biochem J (2020)
Chakraborti S, Balakrishnan D, Trotter AJ, Gittens WH, Yang AWH, Jolma A, Paterson JR, Świątek S, Plewka J, Curtis FA, Bowers LY, Pålsson LO, Hughes TR, Taube M, Kozak M, Heddle JG, Sharples GJ
|
| RgGuinier |
2.1 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
33 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
TRAF-interacting protein with FHA domain-containing protein A dimer, 45 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 100 mM arginine, 5 % glycerol, 10 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, Kumamoto University on 2014 Oct 16
|
Structural analysis of TIFA: Insight into TIFA-dependent signal transduction in innate immunity.
Sci Rep 10(1):5152 (2020)
Nakamura T, Hashikawa C, Okabe K, Yokote Y, Chirifu M, Toma-Fukai S, Nakamura N, Matsuo M, Kamikariya M, Okamoto Y, Gohda J, Akiyama T, Semba K, Ikemizu S, Otsuka M, Inoue JI, Yamagata Y
|
| RgGuinier |
3.1 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Mitochondrial import inner membrane translocase subunit TIM12 monomer, 12 kDa Saccharomyces cerevisiae protein
Mitochondrial import inner membrane translocase subunit TIM9 dimer, 20 kDa Saccharomyces cerevisiae protein
Mitochondrial import inner membrane translocase subunit TIM10 trimer, 31 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
20 mM MES buffer (2-(N-morpholino)ethanesulfonic acid), pH 6.5, 50 mM NaCl Cloning,, pH: 6.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 May 8
|
Architecture and subunit dynamics of the mitochondrial TIM9·10·12 chaperone
(2020)
Weinhäupl K, Wang Y, Hessel A, Brennich M, Lindorff-Larsen K, Schanda P
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
103 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Severe fever with thrombocytopenia syndrome virus L protein - RNA-dependent RNA polymerase monomer, 238 kDa SFTS virus AH12 protein
|
| Buffer: |
50 mM HEPES(NaOH) pH 7.0, 500 mM NaCl, 5% (w/v) glycerol, and 2 mM dithiothreitol, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Sep 13
|
Structural and functional characterization of the Severe fever with thrombocytopenia syndrome virus L protein
(2020)
Vogel D, Thorkelsson S, Quemin E, Meier K, Kouba T, Gogrefe N, Busch C, Reindl S, Günther S, Cusack S, Grünewald K, Rosenthal M
|
| RgGuinier |
4.5 |
nm |
| Dmax |
14.3 |
nm |
| VolumePorod |
483 |
nm3 |
|
|