|
|
|
|
|
| Sample: |
2,4-dichlorophenol 6-monooxygenase hexamer, 399 kDa Streptomyces sp. SCSIO … protein
Flavin adenine dinucleotide hexamer, 5 kDa
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 5 mM DTT, 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at Xenocs BioXolver L with MetalJet, Département de Biochimie, Université de Montréal on 2019 Oct 22
|
Structural analyses of the group A flavin-dependent monooxygenase PieE reveal a sliding FAD cofactor conformation bridging OUT and IN conformations.
J Biol Chem (2020)
Manenda MS, Picard MÈ, Zhang L, Cyr N, Zhu X, Barma J, Pascal JM, Couture M, Zhang C, Shi R
|
| RgGuinier |
4.8 |
nm |
| Dmax |
13.2 |
nm |
| VolumePorod |
624 |
nm3 |
|
|
|
|
|
|
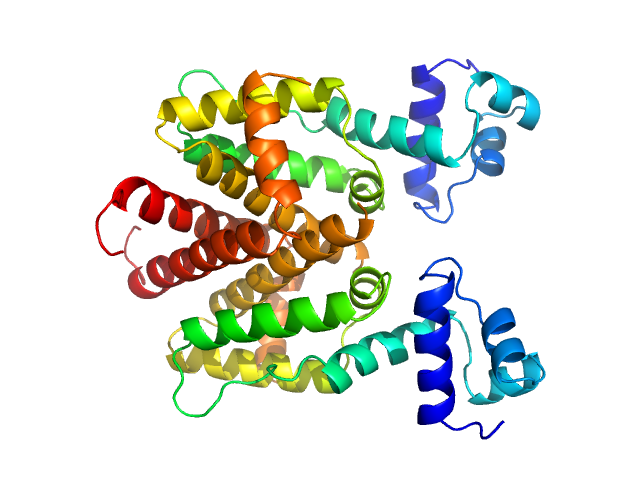
|
| Sample: |
Tetracycline repressor (class D) dimer, 47 kDa Escherichia coli protein
|
| Buffer: |
50 mM Tris/HCl 150 mM NaCl 10 mM MgCl2, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Sep 23
|
Thermodynamics, cooperativity and stability of the tetracycline repressor (TetR) upon tetracycline binding.
Biochim Biophys Acta Proteins Proteom :140404 (2020)
Palm GJ, Buchholz I, Werten S, Girbardt B, Berndt L, Delcea M, Hinrichs W
|
| RgGuinier |
2.6 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
|
|
|
|
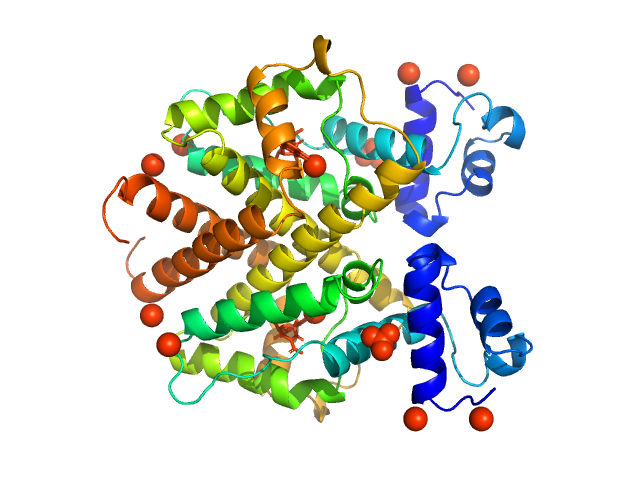
|
| Sample: |
Tetracycline repressor (class D) dimer, 47 kDa Escherichia coli protein
5a,6-anhydrotetracycline dimer, 1 kDa
|
| Buffer: |
50 mM Tris/HCl 150 mM NaCl 10 mM MgCl2, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Sep 23
|
Thermodynamics, cooperativity and stability of the tetracycline repressor (TetR) upon tetracycline binding.
Biochim Biophys Acta Proteins Proteom :140404 (2020)
Palm GJ, Buchholz I, Werten S, Girbardt B, Berndt L, Delcea M, Hinrichs W
|
| RgGuinier |
2.6 |
nm |
| Dmax |
6.8 |
nm |
| VolumePorod |
77 |
nm3 |
|
|
|
|
|
|
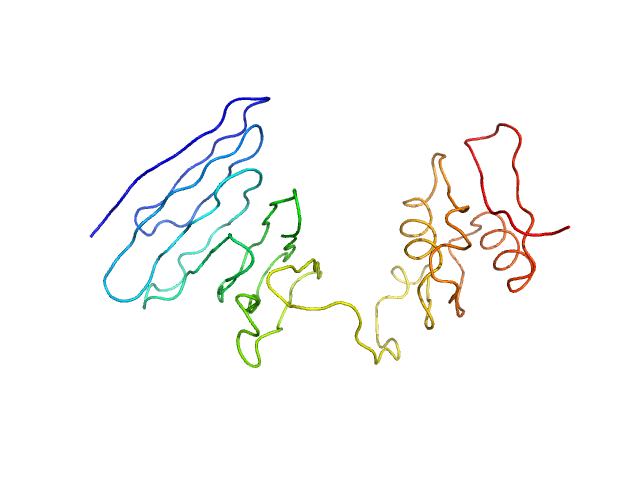
|
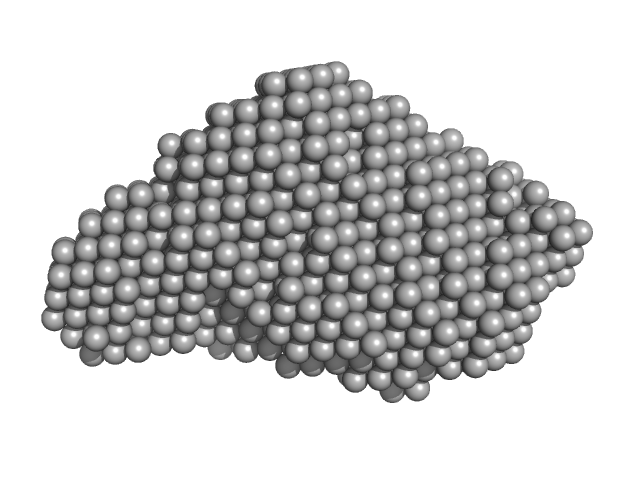
| Sample: |
PupR protein monomer, 24 kDa Pseudomonas putida protein
|
| Buffer: |
25 mM HEPES 400 mM LiCl 10% v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2016 Mar 16
|
Structural basis of cell surface signaling by a conserved sigma regulator in Gram-negative bacteria.
J Biol Chem (2020)
Jensen JL, Jernberg BD, Sinha S, Colbert CL
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
49 |
nm3 |
|
|
|
|
|
|
|
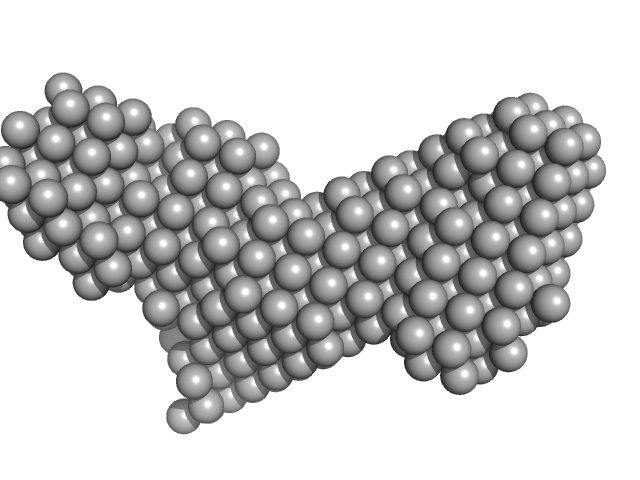
| Sample: |
PupR protein monomer, 24 kDa Pseudomonas putida protein
Ferric-pseudobactin BN7/BN8 receptor monomer, 8 kDa Pseudomonas putida protein
|
| Buffer: |
25 mM HEPES 400 mM LiCl 10% v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2016 Mar 16
|
Structural basis of cell surface signaling by a conserved sigma regulator in Gram-negative bacteria.
J Biol Chem (2020)
Jensen JL, Jernberg BD, Sinha S, Colbert CL
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.7 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
|
|
|
|
|
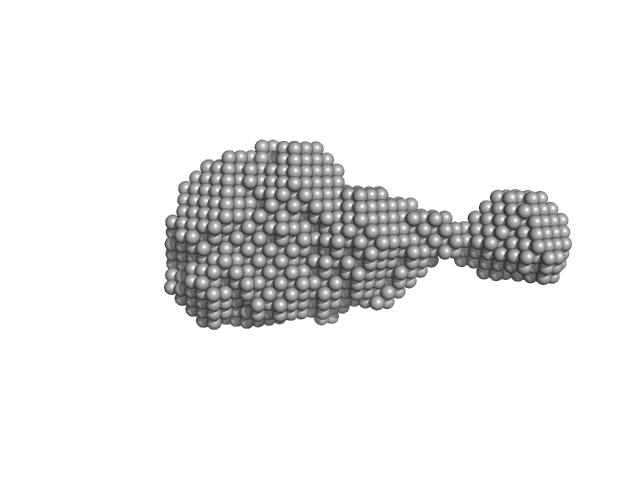
| Sample: |
Histone deacetylase 1 monomer, 55 kDa Homo sapiens protein
Lysine-specific histone demethylase 1A monomer, 93 kDa Homo sapiens protein
REST corepressor 1 monomer, 46 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris/Cl, 50 mM potassium acetate and 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2015 Jan 23
|
Mechanism of Crosstalk between the LSD1 Demethylase and HDAC1 Deacetylase in the CoREST Complex.
Cell Rep 30(8):2699-2711.e8 (2020)
Song Y, Dagil L, Fairall L, Robertson N, Wu M, Ragan TJ, Savva CG, Saleh A, Morone N, Kunze MBA, Jamieson AG, Cole PA, Hansen DF, Schwabe JWR
|
| RgGuinier |
6.0 |
nm |
| Dmax |
15.8 |
nm |
| VolumePorod |
437 |
nm3 |
|
|
|
|
|
|
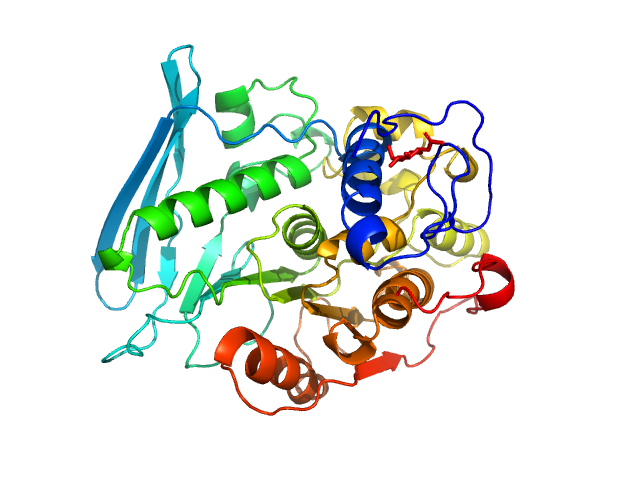
|
| Sample: |
4-O-methyl-glucuronoyl methylesterase (Glucuronoyl esterase) monomer, 51 kDa Cerrena unicolor protein
|
| Buffer: |
20 mM sodium acetate, pH: 5 |
| Experiment: |
SAXS
data collected at Xenocs BioXolver L with GeniX3D, University of Copenhagen, Department of Drug Design and Pharmacology on 2018 Oct 10
|
The structural basis of fungal glucuronoyl esterase activity on natural substrates.
Nat Commun 11(1):1026 (2020)
Ernst HA, Mosbech C, Langkilde AE, Westh P, Meyer AS, Agger JW, Larsen S
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
71 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
4-O-methyl-glucuronoyl methylesterase (Glucuronoyl esterase, truncated) monomer, 43 kDa Cerrena unicolor protein
|
| Buffer: |
20 mM sodium acetate, pH: 5 |
| Experiment: |
SAXS
data collected at Xenocs BioXolver L with GeniX3D, University of Copenhagen, Department of Drug Design and Pharmacology on 2018 Oct 10
|
The structural basis of fungal glucuronoyl esterase activity on natural substrates.
Nat Commun 11(1):1026 (2020)
Ernst HA, Mosbech C, Langkilde AE, Westh P, Meyer AS, Agger JW, Larsen S
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.1 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cation-independent mannose-6-phosphate receptor monomer, 81 kDa Homo sapiens protein
|
| Buffer: |
20 mM imidazole, 150 mM NaCl, 5 mM beta glycerol phosphate, 10 mM MnCl2, pH: 6.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Oct 12
|
Allosteric regulation of lysosomal enzyme recognition by the cation-independent mannose 6-phosphate receptor.
Commun Biol 3(1):498 (2020)
Olson LJ, Misra SK, Ishihara M, Battaile KP, Grant OC, Sood A, Woods RJ, Kim JP, Tiemeyer M, Ren G, Sharp JS, Dahms NM
|
| RgGuinier |
3.7 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
140 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cation-independent mannose-6-phosphate receptor monomer, 81 kDa Homo sapiens protein
|
| Buffer: |
20 mM imidazole, 150 mM NaCl, 5 mM beta glycerol phosphate, 10 mM MnCl2, pH: 6.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Nov 12
|
Allosteric regulation of lysosomal enzyme recognition by the cation-independent mannose 6-phosphate receptor.
Commun Biol 3(1):498 (2020)
Olson LJ, Misra SK, Ishihara M, Battaile KP, Grant OC, Sood A, Woods RJ, Kim JP, Tiemeyer M, Ren G, Sharp JS, Dahms NM
|
| RgGuinier |
3.7 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
147 |
nm3 |
|
|