|
|
|
|
|
| Sample: |
Cation-independent mannose-6-phosphate receptor monomer, 81 kDa Homo sapiens protein
|
| Buffer: |
20 mM imidazole, 150 mM NaCl, 5 mM beta glycerol phosphate, 10 mM MnCl2, pH: 6.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Nov 12
|
Allosteric regulation of lysosomal enzyme recognition by the cation-independent mannose 6-phosphate receptor.
Commun Biol 3(1):498 (2020)
Olson LJ, Misra SK, Ishihara M, Battaile KP, Grant OC, Sood A, Woods RJ, Kim JP, Tiemeyer M, Ren G, Sharp JS, Dahms NM
|
| RgGuinier |
3.7 |
nm |
| Dmax |
13.3 |
nm |
| VolumePorod |
150 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Palmitoyl-protein thioesterase 1 monomer, 31 kDa Homo sapiens protein
|
| Buffer: |
20 mM imidazole, 150 mM NaCl, 5 mM beta glycerol phosphate, 10 mM MnCl2, pH: 6.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Nov 12
|
Allosteric regulation of lysosomal enzyme recognition by the cation-independent mannose 6-phosphate receptor.
Commun Biol 3(1):498 (2020)
Olson LJ, Misra SK, Ishihara M, Battaile KP, Grant OC, Sood A, Woods RJ, Kim JP, Tiemeyer M, Ren G, Sharp JS, Dahms NM
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
54 |
nm3 |
|
|
|
|
|
|
|
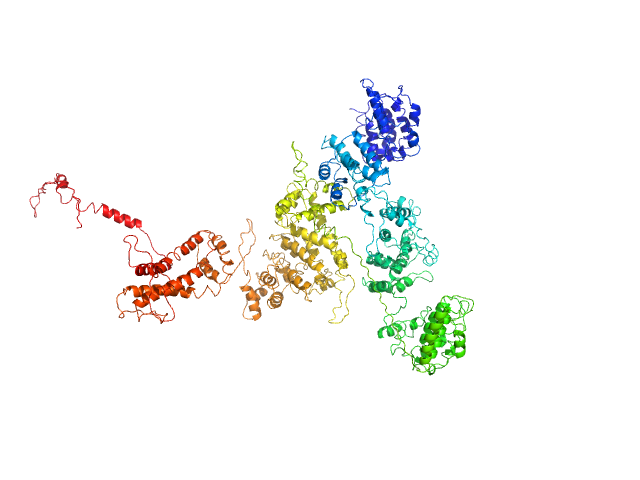
| Sample: |
Cation-independent mannose-6-phosphate receptor monomer, 81 kDa Homo sapiens protein
Palmitoyl-protein thioesterase 1 monomer, 31 kDa Homo sapiens protein
|
| Buffer: |
20 mM imidazole, 150 mM NaCl, 5 mM beta glycerol phosphate, 10 mM MnCl2, pH: 6.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Nov 12
|
Allosteric regulation of lysosomal enzyme recognition by the cation-independent mannose 6-phosphate receptor.
Commun Biol 3(1):498 (2020)
Olson LJ, Misra SK, Ishihara M, Battaile KP, Grant OC, Sood A, Woods RJ, Kim JP, Tiemeyer M, Ren G, Sharp JS, Dahms NM
|
| RgGuinier |
4.9 |
nm |
| Dmax |
19.3 |
nm |
| VolumePorod |
258 |
nm3 |
|
|
|
|
|
|
|
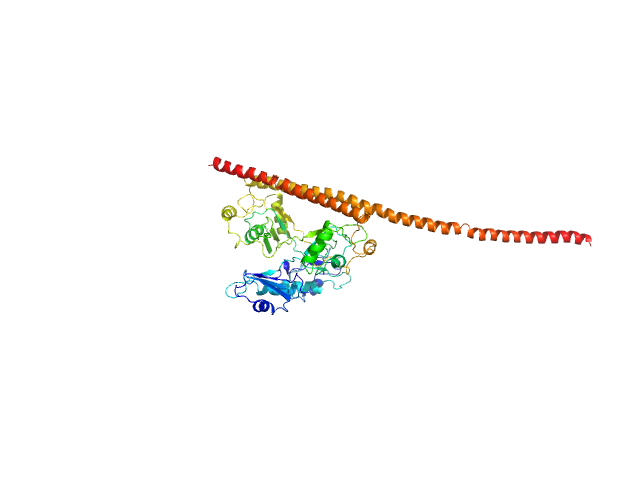
| Sample: |
Macrophage mannose receptor 1 dimer, 315 kDa Mouse myeloma cell … protein
|
| Buffer: |
50mM Hepes, 100mM NaCl, 1mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2016 Apr 15
|
Mannose receptor (CD206) activation in tumor-associated macrophages enhances adaptive and innate antitumor immune responses.
Sci Transl Med 12(530) (2020)
Jaynes JM, Sable R, Ronzetti M, Bautista W, Knotts Z, Abisoye-Ogunniyan A, Li D, Calvo R, Dashnyam M, Singh A, Guerin T, White J, Ravichandran S, Kumar P, Talsania K, Chen V, Ghebremedhin A, Karanam B, Bin Salam A, Amin R, Odzorig T, Aiken T, Nguyen V, Bian Y, Zarif JC, de Groot AE, Mehta M, Fan L, Hu X, Simeonov A, Pate N, Abu-Asab M, Ferrer M, Southall N, Ock CY, Zhao Y, Lopez H, Kozlov S, de Val N, Yates CC, Baljinnyam B, Marugan J, Rudloff U
|
| RgGuinier |
7.9 |
nm |
| Dmax |
30.1 |
nm |
| VolumePorod |
584 |
nm3 |
|
|
|
|
|
|
|
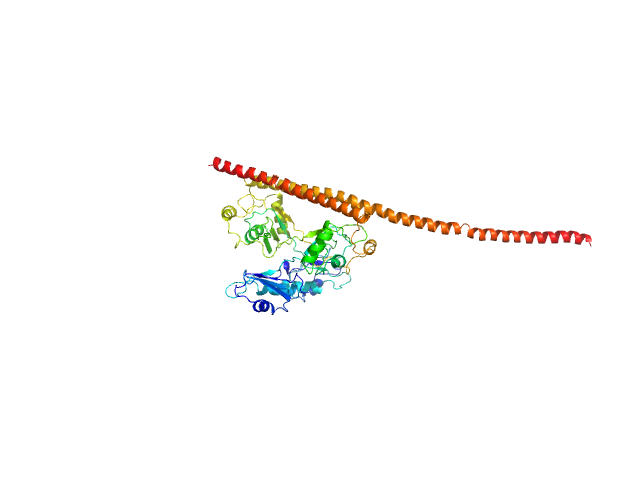
| Sample: |
Non-POU domain-containing octamer-binding protein monomer, 30 kDa Homo sapiens protein
Splicing factor, proline- and glutamine-rich monomer, 38 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 250 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Apr 5
|
Structural basis of the zinc-induced cytoplasmic aggregation of the RNA-binding protein SFPQ
Nucleic Acids Research 48(6):3356-3365 (2020)
Huang J, Ringuet M, Whitten A, Caria S, Lim Y, Badhan R, Anggono V, Lee M
|
| RgGuinier |
4.0 |
nm |
| Dmax |
16.5 |
nm |
| VolumePorod |
90 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Splicing factor, proline- and glutamine-rich monomer, 38 kDa Homo sapiens protein
Non-POU domain-containing octamer-binding protein monomer, 30 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 250 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Nov 8
|
Structural basis of the zinc-induced cytoplasmic aggregation of the RNA-binding protein SFPQ
Nucleic Acids Research 48(6):3356-3365 (2020)
Huang J, Ringuet M, Whitten A, Caria S, Lim Y, Badhan R, Anggono V, Lee M
|
| RgGuinier |
3.6 |
nm |
| Dmax |
16.5 |
nm |
| VolumePorod |
80 |
nm3 |
|
|
|
|
|
|
|
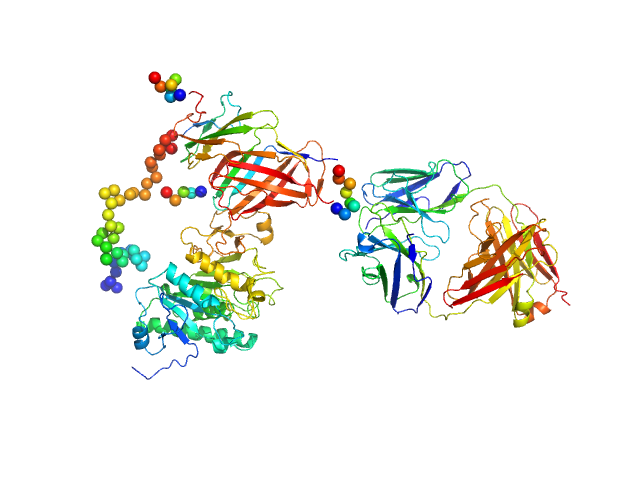
| Sample: |
Lipoprotein lipase monomer, 50 kDa Homo sapiens protein
Glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1 monomer, 15 kDa Homo sapiens protein
Monoclonal Antibody Fragment 5D2 monomer, 47 kDa Mus musculus protein
|
| Buffer: |
10 mM Tris, 150 mM NaCl, 4 mM CaCL2, 10% (v/v) Glycerol, 0.05% 0.8mM CHAPS, ,0.05 % (v/v) NaN3, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Dec 14
|
Unfolding of monomeric lipoprotein lipase by ANGPTL4: Insight into the regulation of plasma triglyceride metabolism
Proceedings of the National Academy of Sciences :201920202 (2020)
Kristensen K, Leth-Espensen K, Mertens H, Birrane G, Meiyappan M, Olivecrona G, Jørgensen T, Young S, Ploug M
|
| RgGuinier |
4.9 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
187 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Latency associated peptide dimer, 58 kDa Homo sapiens protein
|
| Buffer: |
phosphate buffered saline 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Apr 20
|
Structural insights into conformational switching in latency-associated peptide between transforming growth factor β-1 bound and unbound states
IUCrJ 7(2) (2020)
Stachowski T, Snell M, Snell E
|
| RgGuinier |
3.5 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
129 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Lysozyme amyloid fibril, 14 kDa Gallus gallus protein
|
| Buffer: |
0.2 M glycine-HCl, 80 mM NaCl, pH: 2.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Sep 5
|
Effect of the concentration of protein and nanoparticles on the structure of biohybrid nanocomposites.
Biopolymers 111(2):e23342 (2020)
Majorošová J, Schroer MA, Tomašovičová N, Batková M, Hu PS, Kubovčíková M, Svergun DI, Kopčanský P
|
| RgGuinier |
30.2 |
nm |
| Dmax |
120.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Lysozyme amyloid fibril, 1 kDa Gallus gallus protein
Fe3O4 nanoparticles; radius 5.6 nm (AFM based) monomer, 1 kDa
|
| Buffer: |
0.2 M glycine-HCl, 80 mM NaCl, pH: 2.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Sep 8
|
Effect of the concentration of protein and nanoparticles on the structure of biohybrid nanocomposites.
Biopolymers 111(2):e23342 (2020)
Majorošová J, Schroer MA, Tomašovičová N, Batková M, Hu PS, Kubovčíková M, Svergun DI, Kopčanský P
|
| RgGuinier |
29.4 |
nm |
| Dmax |
80.0 |
nm |
|
|