|
|
|
|
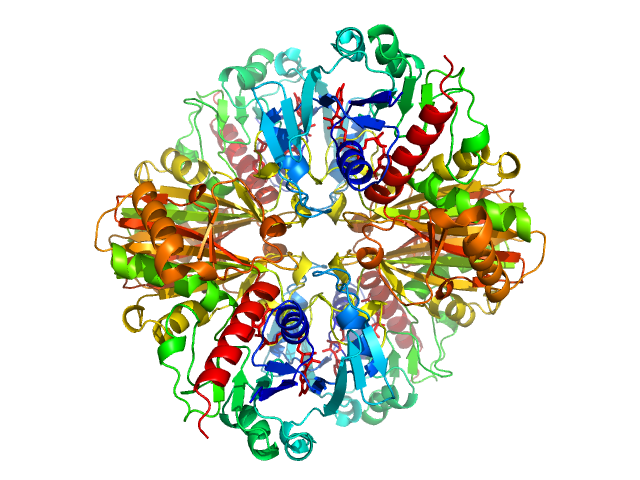
|
| Sample: |
Glyceraldehyde-3-phosphate dehydrogenase tetramer, 148 kDa Chlamydomonas reinhardtii protein
|
| Buffer: |
30 mM Tris, 4 mM EDTA, 100 µM NAD, 5 mM free cysteine, pH: 7.9
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2011 Feb 7
|
Cryptic Disorder Out of Disorder: Encounter between Conditionally Disordered CP12 and Glyceraldehyde-3-Phosphate Dehydrogenase.
J Mol Biol 430(8):1218-1234 (2018)
Launay H, Barré P, Puppo C, Zhang Y, Maneville S, Gontero B, Receveur-Bréchot V
|
| RgGuinier |
3.2 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
206 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Calvin cycle protein CP12, chloroplastic monomer, 11 kDa Chlamydomonas reinhardtii protein
Glyceraldehyde-3-phosphate dehydrogenase tetramer, 148 kDa Chlamydomonas reinhardtii protein
|
| Buffer: |
30 mM Tris, 4 mM EDTA, 100 µM NAD, 5 mM free cysteine, pH: 7.9
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2010 Dec 8
|
Cryptic Disorder Out of Disorder: Encounter between Conditionally Disordered CP12 and Glyceraldehyde-3-Phosphate Dehydrogenase.
J Mol Biol 430(8):1218-1234 (2018)
Launay H, Barré P, Puppo C, Zhang Y, Maneville S, Gontero B, Receveur-Bréchot V
|
| RgGuinier |
3.8 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
251 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Protein sex-lethal monomer, 20 kDa Drosophila melanogaster protein
|
| Buffer: |
10 mM KP, 50 mM NaCl, 10 mM DTT, pH: 6
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Oct 16
|
A General Small-Angle X-ray Scattering-Based Screening Protocol Validated for Protein-RNA Interactions.
ACS Comb Sci 20(4):197-202 (2018)
Chen PC, Masiewicz P, Rybin V, Svergun D, Hennig J
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.9 |
nm |
| VolumePorod |
26 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Protein sex-lethal mutant monomer, 20 kDa Drosophila melanogaster protein
|
| Buffer: |
10 mM KP, 50 mM NaCl, 10 mM DTT, pH: 6
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Feb 2
|
A General Small-Angle X-ray Scattering-Based Screening Protocol Validated for Protein-RNA Interactions.
ACS Comb Sci 20(4):197-202 (2018)
Chen PC, Masiewicz P, Rybin V, Svergun D, Hennig J
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Protein sex-lethal mutant monomer, 20 kDa Drosophila melanogaster protein
RNA decaneucleotide U8GU monomer, 3 kDa synthetic construct RNA
|
| Buffer: |
50% dilution of protein buffer {10 mM KP, 50 mM NaCl, 10 mM DTT pH 6} with {milliQ-water pH 7} suspended RNA, pH: 6
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Feb 3
|
A General Small-Angle X-ray Scattering-Based Screening Protocol Validated for Protein-RNA Interactions.
ACS Comb Sci 20(4):197-202 (2018)
Chen PC, Masiewicz P, Rybin V, Svergun D, Hennig J
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
34 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Protein sex-lethal mutant dimer, 41 kDa Drosophila melanogaster protein
RNA decaneucleotide UGU8 dimer, 6 kDa synthetic construct RNA
|
| Buffer: |
50% dilution of protein buffer {10 mM KP, 50 mM NaCl, 10 mM DTT pH 6} with {milliQ-water pH 7} suspended RNA, pH: 6
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Apr 12
|
A General Small-Angle X-ray Scattering-Based Screening Protocol Validated for Protein-RNA Interactions.
ACS Comb Sci 20(4):197-202 (2018)
Chen PC, Masiewicz P, Rybin V, Svergun D, Hennig J
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|
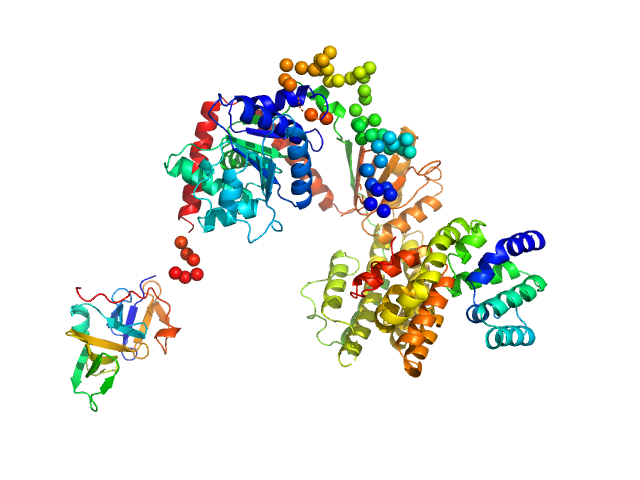
|
| Sample: |
Probable ATP-dependent RNA helicase DDX58 monomer, 108 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 2.5 mM MgCl2, 10% glycerol and 1mM DTT, pH: 7.4
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2012 Apr 6
|
Combined roles of ATP and small hairpin RNA in the activation of RIG-I revealed by solution-based analysis.
Nucleic Acids Res 46(6):3169-3186 (2018)
Shah N, Beckham SA, Wilce JA, Wilce MCJ
|
| RgGuinier |
4.3 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
186 |
nm3 |
|
|
|
|
|
|
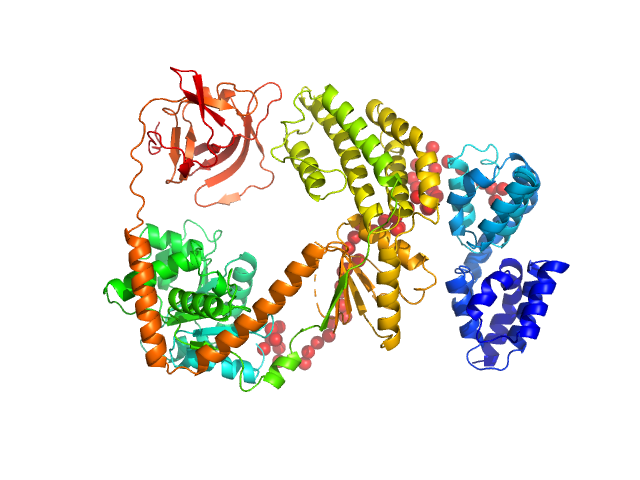
|
| Sample: |
Probable ATP-dependent RNA helicase DDX58 monomer, 108 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 2.5 mM MgCl2, 10% glycerol and 1mM DTT, 2mM ADP-AlFx, pH: 7.4
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Nov 20
|
Combined roles of ATP and small hairpin RNA in the activation of RIG-I revealed by solution-based analysis.
Nucleic Acids Res 46(6):3169-3186 (2018)
Shah N, Beckham SA, Wilce JA, Wilce MCJ
|
| RgGuinier |
4.2 |
nm |
| Dmax |
15.6 |
nm |
| VolumePorod |
190 |
nm3 |
|
|
|
|
|
|
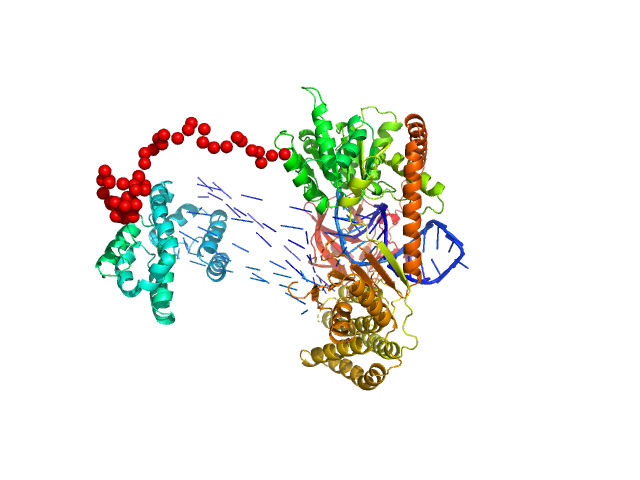
|
| Sample: |
Probable ATP-dependent RNA helicase DDX58 monomer, 108 kDa Homo sapiens protein
5´ppp 10mer hairpin dsRNA monomer, 8 kDa RNA
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 2.5 mM MgCl2, 10% glycerol and 1mM DTT, pH: 7.4
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 May 29
|
Combined roles of ATP and small hairpin RNA in the activation of RIG-I revealed by solution-based analysis.
Nucleic Acids Res 46(6):3169-3186 (2018)
Shah N, Beckham SA, Wilce JA, Wilce MCJ
|
| RgGuinier |
4.1 |
nm |
| Dmax |
16.1 |
nm |
| VolumePorod |
160 |
nm3 |
|
|
|
|
|
|
|
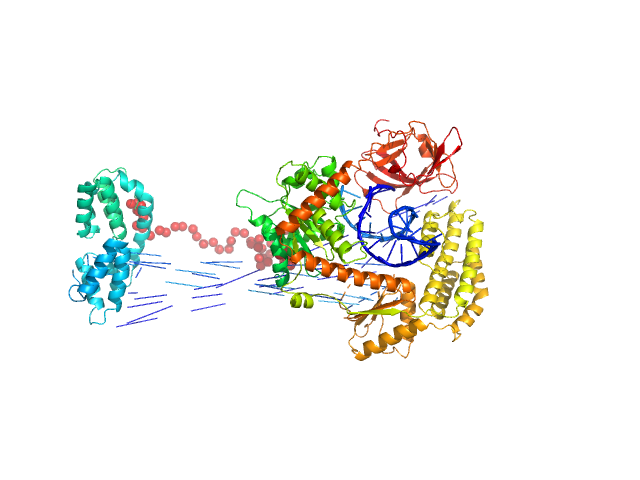
| Sample: |
Probable ATP-dependent RNA helicase DDX58 monomer, 108 kDa Homo sapiens protein
5´ppp 10mer hairpin dsRNA monomer, 8 kDa RNA
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 2.5 mM MgCl2, 10% glycerol and 1mM DTT, 0.5 mM AMP-PNP, pH: 7.4
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 May 28
|
Combined roles of ATP and small hairpin RNA in the activation of RIG-I revealed by solution-based analysis.
Nucleic Acids Res 46(6):3169-3186 (2018)
Shah N, Beckham SA, Wilce JA, Wilce MCJ
|
| RgGuinier |
4.2 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
163 |
nm3 |
|
|