| MWI(0) | 100 | kDa |
| MWexpected | 169 | kDa |
| VPorod | 166 | nm3 |
|
log I(s)
1.75×10-1
1.75×10-2
1.75×10-3
1.75×10-4
|
 s, nm-1
s, nm-1
|
|
|
|
|
|

|
|
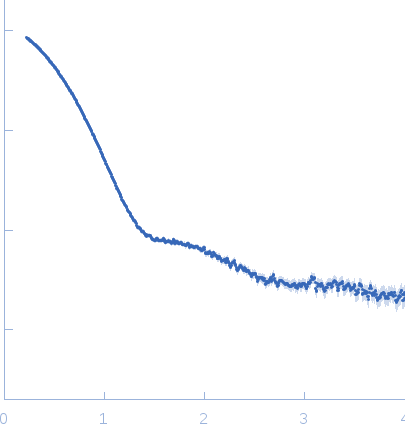
Synchrotron SAXS
data from solutions of
EcPaaA2-HisEcParE2 construct
in
50 mM Tris-HCl 500 mM NaCl, pH 7.5
were collected
on the
SWING beam line
at the SOLEIL storage ring
(Saint-Aubin, France)
using a AVIEX PCCD170170 detector
at a sample-detector distance of 1.8 m and
at a wavelength of λ = 0.1 nm
(I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle).
Solute concentrations ranging between 1 and 10 mg/ml were measured
at 10°C.
750 successive
0.750 second frames were collected.
The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
The low angle data collected at lower concentration were merged with the highest concentration high angle data to yield the final composite scattering curve.
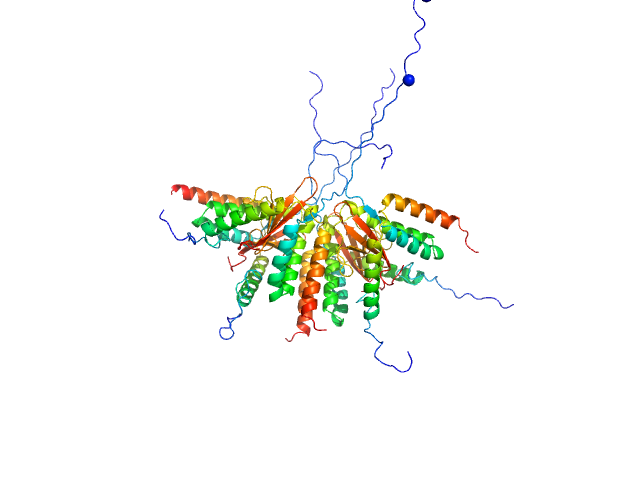
Use of SAXS to validate the oligomer state of the obtained crystal structure and to model missing afinity tags |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||