|
|
|
|
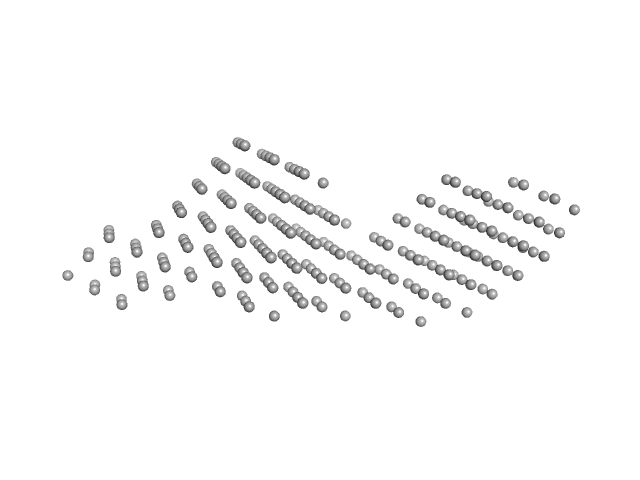
|
| Sample: |
Protein TrbB (GST-fusion) dimer, 93 kDa Escherichia coli (strain … protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 5% glycerol, 0.05% NP40, pH: 7
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Feb 23
|
Structural insights into the disulfide isomerase and chaperone activity of TrbB of the F plasmid Type IV Secretion System
Arnold Apostol
|
| RgGuinier |
4.0 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
153 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDQZ7_fit2_model1.png)
|
| Sample: |
Ubiquitin carboxyl-terminal hydrolase 14 monomer, 56 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Oct 18
|
Transient interdomain interactions in free USP14 shape its conformational ensemble.
Protein Sci 33(5):e4975 (2024)
Salomonsson J, Wallner B, Sjöstrand L, D'Arcy P, Sunnerhagen M, Ahlner A
|
| RgGuinier |
3.1 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
111 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Ubiquitin carboxyl-terminal hydrolase 14 monomer, 46 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 5
|
Transient interdomain interactions in free USP14 shape its conformational ensemble.
Protein Sci 33(5):e4975 (2024)
Salomonsson J, Wallner B, Sjöstrand L, D'Arcy P, Sunnerhagen M, Ahlner A
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
88 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
beta-glucosidase dimer, 187 kDa Aspergillus niger protein
|
| Buffer: |
50 mM sodium acetate, pH: 5
|
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2024 Feb 15
|
Aspergillus niger beta-glucosidase (bgl1)
Estella Yee
|
| RgGuinier |
4.2 |
nm |
| Dmax |
13.9 |
nm |
| VolumePorod |
306 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Isoform A1B1 of Teneurin-3 dimer, 545 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.8
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Sep 10
|
Alternative splicing controls teneurin-3 compact dimer formation for neuronal recognition
Nature Communications 15(1) (2024)
Gogou C, Beugelink J, Frias C, Kresik L, Jaroszynska N, Drescher U, Janssen B, Hindges R, Meijer D
|
| RgGuinier |
8.9 |
nm |
| Dmax |
37.0 |
nm |
| VolumePorod |
1059 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Isoform A1B1 of Teneurin-3 dimer, 545 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM CaCl2, pH: 7.8
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Sep 10
|
Alternative splicing controls teneurin-3 compact dimer formation for neuronal recognition
Nature Communications 15(1) (2024)
Gogou C, Beugelink J, Frias C, Kresik L, Jaroszynska N, Drescher U, Janssen B, Hindges R, Meijer D
|
| RgGuinier |
7.9 |
nm |
| Dmax |
33.0 |
nm |
| VolumePorod |
1133 |
nm3 |
|
|
|
|
|
|
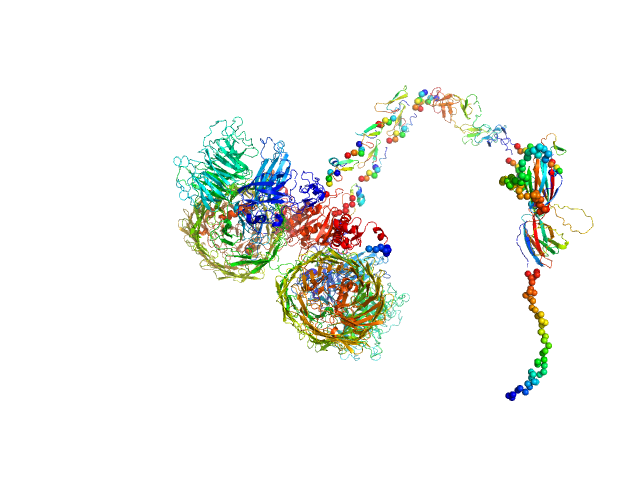
|
| Sample: |
Isoform A1B1 of Teneurin-3 dimer, 545 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM CaCl2, pH: 7.8
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Sep 10
|
Alternative splicing controls teneurin-3 compact dimer formation for neuronal recognition
Nature Communications 15(1) (2024)
Gogou C, Beugelink J, Frias C, Kresik L, Jaroszynska N, Drescher U, Janssen B, Hindges R, Meijer D
|
| RgGuinier |
7.9 |
nm |
| Dmax |
32.0 |
nm |
| VolumePorod |
1129 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Isoform A1B1 of Teneurin-3 dimer, 545 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM CaCl2, pH: 7.8
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Sep 10
|
Alternative splicing controls teneurin-3 compact dimer formation for neuronal recognition
Nature Communications 15(1) (2024)
Gogou C, Beugelink J, Frias C, Kresik L, Jaroszynska N, Drescher U, Janssen B, Hindges R, Meijer D
|
| RgGuinier |
7.9 |
nm |
| Dmax |
33.0 |
nm |
| VolumePorod |
1129 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Isoform A1B1 of Teneurin-3 dimer, 545 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM EDTA, pH: 7.8
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Sep 10
|
Alternative splicing controls teneurin-3 compact dimer formation for neuronal recognition
Nature Communications 15(1) (2024)
Gogou C, Beugelink J, Frias C, Kresik L, Jaroszynska N, Drescher U, Janssen B, Hindges R, Meijer D
|
| RgGuinier |
8.8 |
nm |
| Dmax |
35.0 |
nm |
| VolumePorod |
1035 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Isoform A0B0 of Teneurin-3 dimer, 526 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.8
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Sep 10
|
Alternative splicing controls teneurin-3 compact dimer formation for neuronal recognition
Nature Communications 15(1) (2024)
Gogou C, Beugelink J, Frias C, Kresik L, Jaroszynska N, Drescher U, Janssen B, Hindges R, Meijer D
|
| RgGuinier |
9.4 |
nm |
| Dmax |
39.0 |
nm |
| VolumePorod |
1101 |
nm3 |
|
|