|
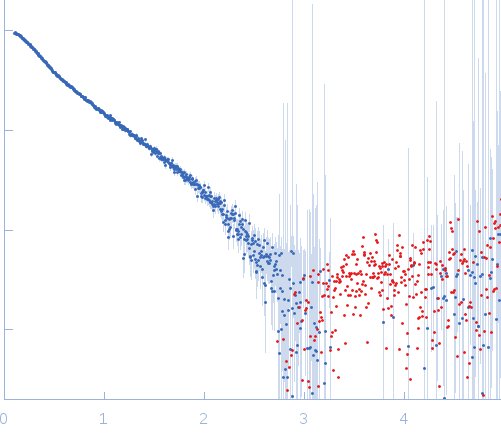
Synchrotron SAXS
data from solutions of
kLBS1-2 DNA
in
Tris, pH 7.6
were collected
on the
BM29 beam line
at the ESRF storage ring
(Grenoble, France)
using a Pilatus 1M detector
at a sample-detector distance of 2.4 m and
at a wavelength of λ = 0.93 nm
(I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle).
One solute concentration of 0.50 mg/ml was measured.
The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
Study of oligomeric states of mLANA and kLANA and their complexes with DNA
|
|
kLBS1-2 DNA
|
| Mol. type |
|
DNA |
| Organism |
|
unidentified herpesvirus |
| Olig. state |
|
Monomer |
| Mon. MW |
|
24 kDa |
|
 s, nm-1
s, nm-1
