|
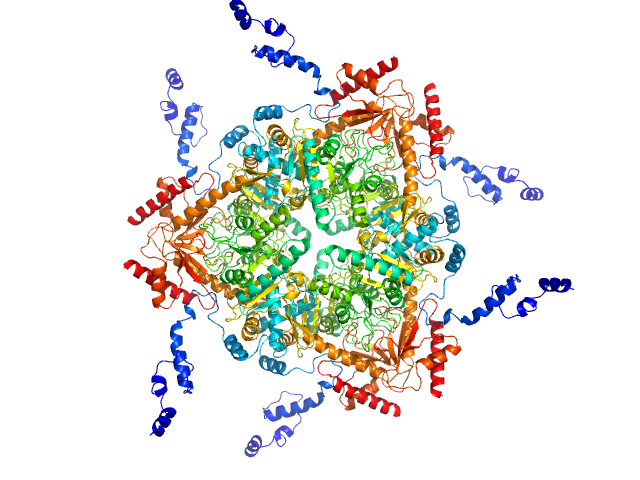
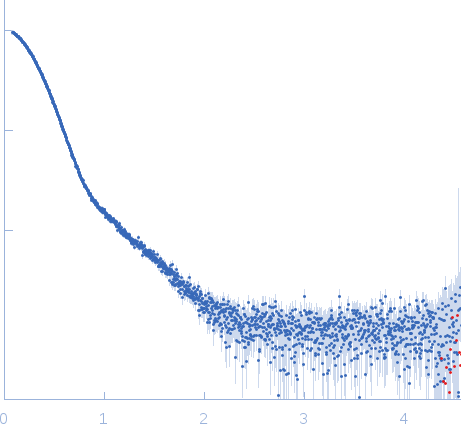
Synchrotron SAXS data from solutions of Glutamate decarboxylase alpha (GadA) from E. coli in 50 mM Tris, pH 7.5 were collected on the P12 beam line of Petra-III (Hamburg, Germany) using a Pilatus 2M detector (I(s) vs s; s = 4π sin θ/λ, where 2θ is the scattering angle and λ=0.124 nm). Different solute concentrations in the range 1.8-8.5 mg/ml were measured using an exposure time of 1 s (recorded as 20 x 0.050 s frames). The data were normalized to the intensity of the transmitted beam and radially averaged and the scattering from the matched solvent-blank was subtracted. The data presented here are from a single concentration scattering curve (8.5 mg/ml).
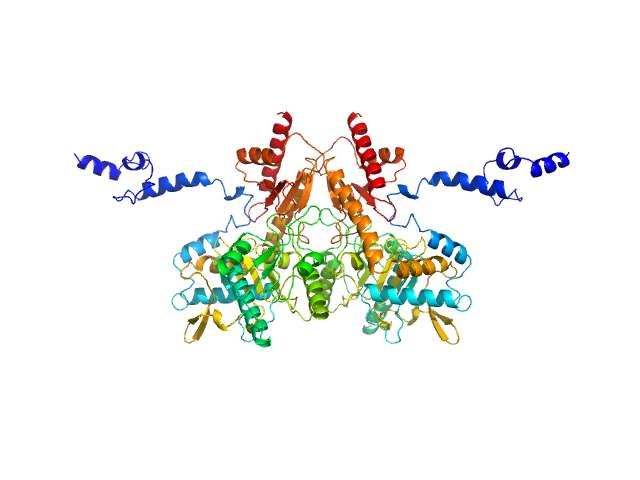
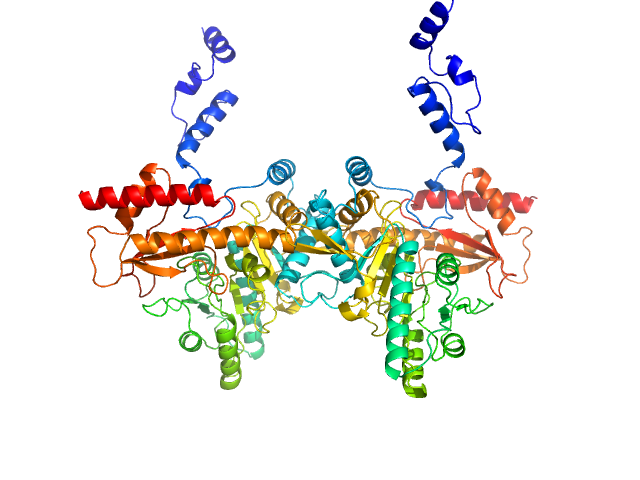
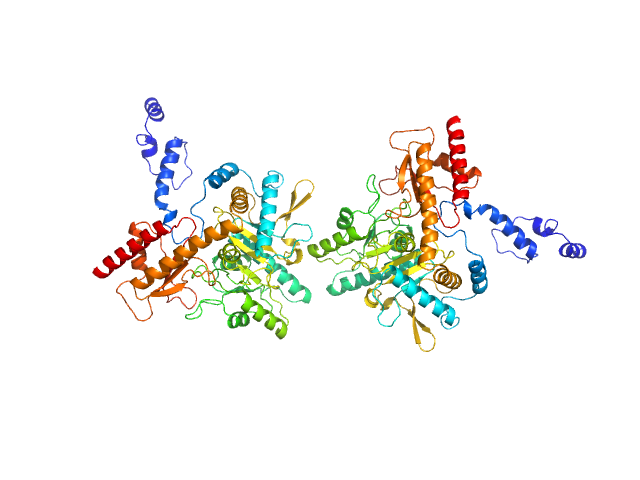
Under the experimental conditions described above, GadA exists as a mixture in solution of likely hexamers (volume fraction approximately 60%) and disassociated dimers (40%). The models displayed for this entry and associated fit are derived from SASREFMX modelling in P32 symmetry. The final fit to the SAXS data of the mixture was determined using OLIGOMER. The specific volume fraction estimates of the hexamer and three dimers are included in the full entry zip archive.
|
|
 s, nm-1
s, nm-1