|
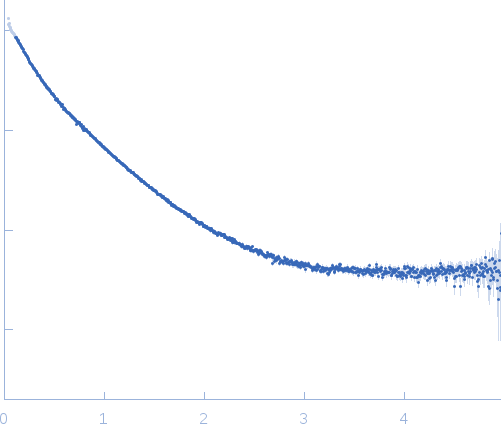
Synchrotron SAXS
data from solutions of
Compound 1:BRD4 (1:1)
in
20mM Hepes, 100mM NaCl, 1mM Tris(2-carboxyethyl)phosphine hydrochloride, pH 7.4
were collected
on the
BM29 beam line
at the ESRF storage ring
(Grenoble, France)
using a Pilatus 1M detector
at a sample-detector distance of 2.9 m and
at a wavelength of λ = 0.0992 nm
(I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle).
Solute concentrations ranging between 1 and 10 mg/ml were measured
.
50 successive
0.200 second frames were collected.
The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
The low angle data collected at lower concentrations were extrapolated to infinite dilution and merged with the higher concentration data to yield the final composite scattering curve.
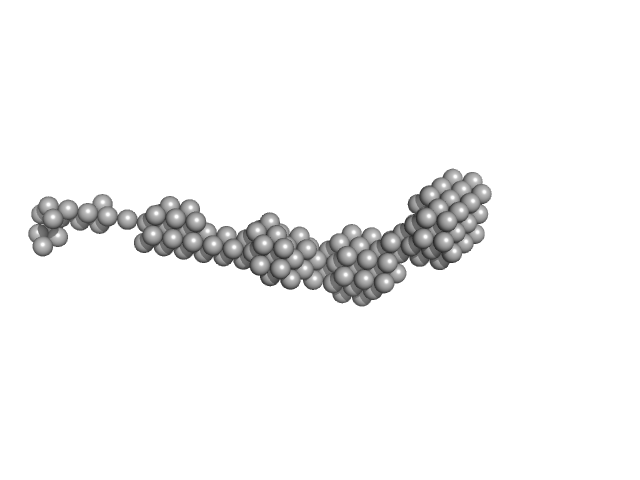
A pool of 10000 models comprising two rigid bromodomains (residues 42-168, PDB ID: 2oss and 349-458, PDB ID: 2yem) connected by a flexible linker, N-terminal tag and flexible N- and C-termini were generated by the program RANCH (EOM). The scattering from each model from the pool was calculated with the program CRYSOL. For the 100 models best fitting the experimental scattering the histograms of the distances between the centres of the two bromodomains were computed; the average distance was 13±2 nm, the average Rg was 6.6±0.7 nm.
|
|
 s, nm-1
s, nm-1