|
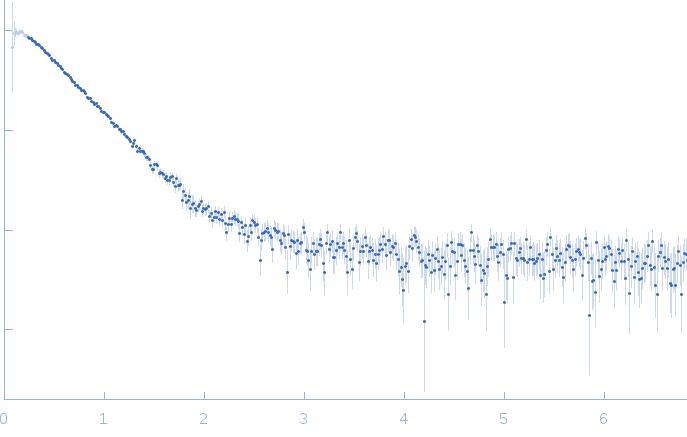
SAXS data collected from solutions of the GDP bound form of the C-terminal deletion mutant of ObgE from E.coli (ObgE_340 with GDP) in 20 mM HEPES, 300 mM NaCl, 250 mM imidazole, 5 mM MgCl2, 2 mM DTT, 400 µM GDP, pH 7.5, were collected at the Rigaku BioSAXS-2000 instrument
using a CCD detector at a sample-detector distance of 1.0 m and at a wavelength of λ = 0.154 nm (I(s) vs s, where s = 4πsinθ/λ and 2θ is the scattering angle). Data collection was performed at at 10°C at four different concentrations of 1, 2, 4 and 8 mg/ml with 30 min exposure each intermittent by reference buffer. Rigaku SAXSLab was used for data reduction.
The sample protein includes an N-terminal poly-histidine tag. Data was merged using PRIMUS from ATSAS package.
|
|
 s, nm-1
s, nm-1
