|
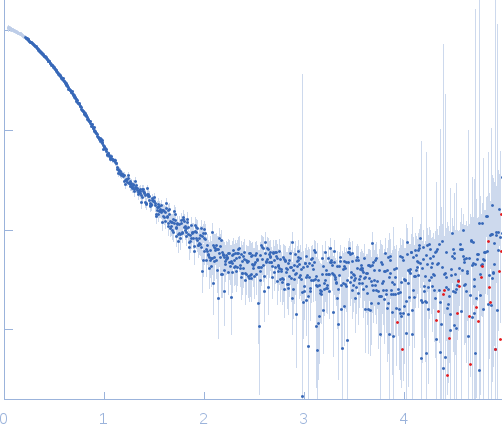
X-ray synchrotron radiation scattering data from solution of PtAUREO1a bZIP-LOV module in 0.5X TBE (50 mM Tris pH 8, 50 mM boric acid, 1 mM EDTA) buffer was collected on the BM29 camera on the storage ring ESRF (Grenoble, France) using a 2D Photon counting Pilatus 1M-W pixel detector (s = 4π sin θ/λ, where 2θ is the scattering angle). Ten successive 20 second frames were collected. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
Solution scattering data of bZIP-LOV module in the light state was performed at 1.25mg/mL concentration at 4 degrees centigrade. The modelling analysis was performed using the ATSAS package. The displayed model is an averaged and volume corrected dummy atom representation (DAMFILT). For further sequence information refer to the Joint Genome Institute (JGI) entry JGI-49116.
|
|
 s, nm-1
s, nm-1
