|
Synchrotron SAXS data from solutions of Class I fructose-1,6-bisphosphate aldolase (FbaB) from E. coli in 50 mM Tris pH 7.5 were collected on the P12 beam line of Petra-III (Hamburg, Germany) using a Pilatus 2M detector (I(s) vs s; s = 4π sin θ/λ, where 2θ is the scattering angle and λ=0.124 nm). Different solute concentrations in the range 2.6-13.30 mg/ml were measured using an exposure time of 1 s (recorded as 20 x 0.050 s frames). The data were normalized to the intensity of the transmitted beam and radially averaged and the scattering from the matched solvent-blank was subtracted. The data presented here are from a single concentration scattering curve (9.3 mg/ml).
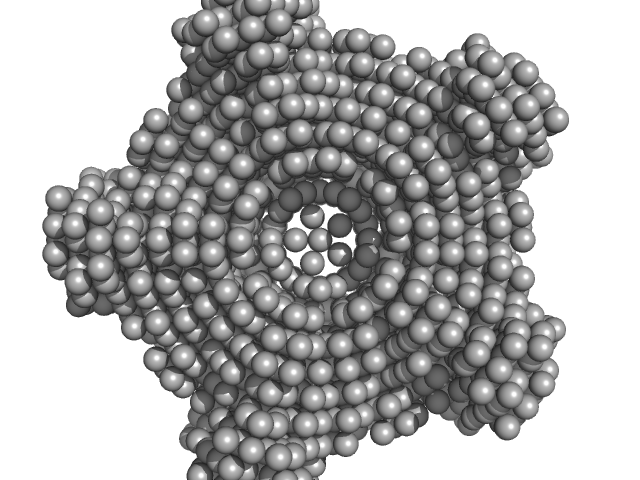
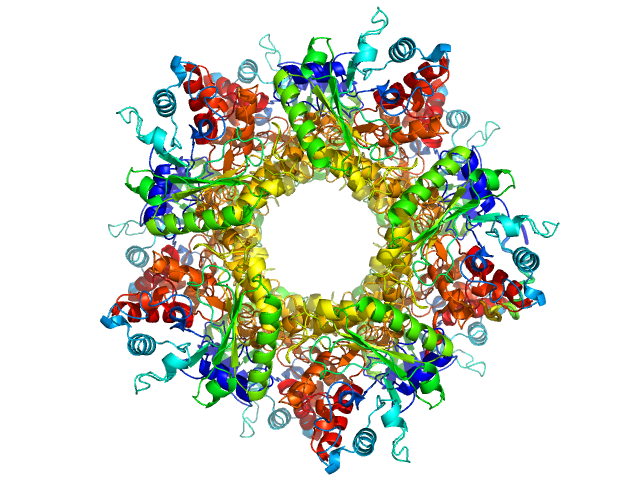
The models displayed above are, from top to bottom: 1) An individual DAMMIN dummy atom model and; 2) CORAL rigid body model with the associated CRYSOL fit to the data. All of the individual DAMMIN models and associated fits can be found in the zip archive for this entry along with the CORAL modelling results.
|
|
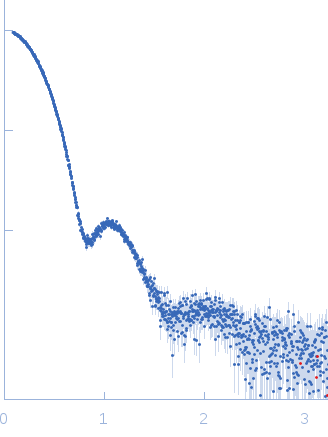 s, nm-1
s, nm-1