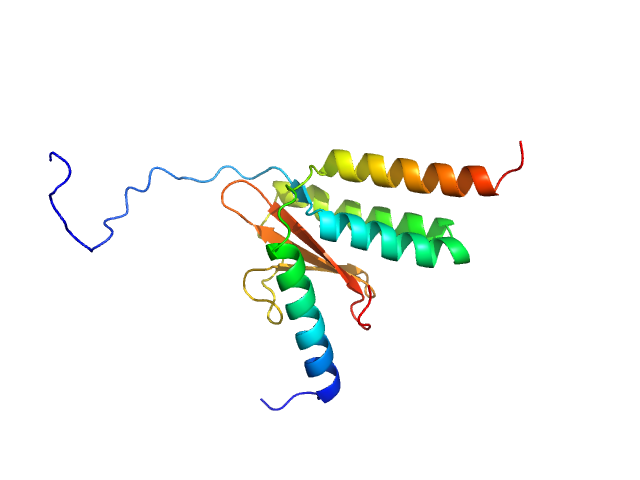
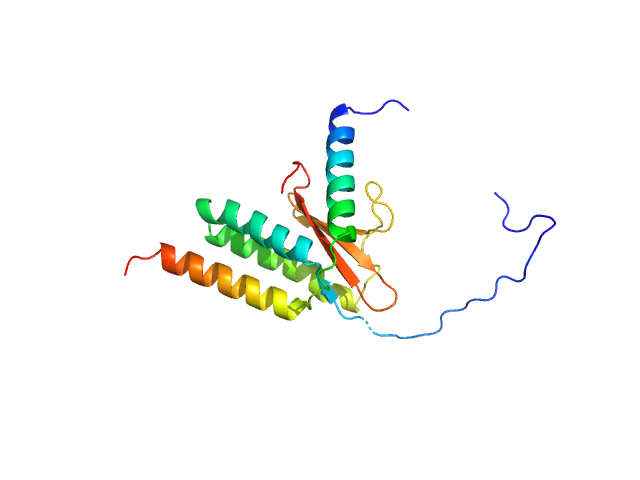
|
Synchrotron SAXS data from EcPaaA2_13-63-EcParE2 were collected at the SWING beamline (SOLEIL synchrotron, Gif-sur-Yvette, France) in HPLC mode using a CCD AVIEX detector at a sample-detector distance of 1.8 m and at a wavelength of λ = 0.103 nm (s = 4π sin θ/λ, where 2θ is the scattering angle). Sample was loaded onto a Shodex KW402.5-4F, which was pre-equilibrated with running buffer (50 mM Tris-HCl, 500 mM NaCl, pH 7.5) for at least one column volume. The flow rate was set to 0.2 ml/min. During each HPLC run, 250 frames were collected after 15 min (0.62 CV) using 750 ms per frame and 1500 ms dead time. After radial averaging and buffer subtraction of the data frames using FOXTROT, the scattering curves were processed and analyzed with DATASW.
Concentration min = UNKNOWN
|
|
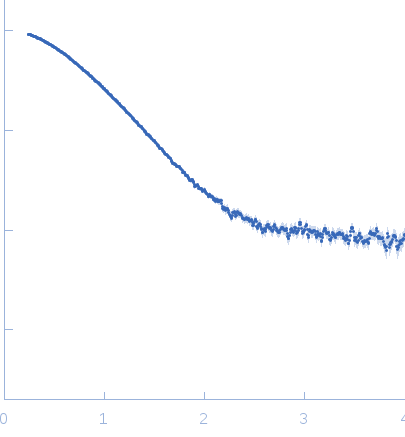 s, nm-1
s, nm-1
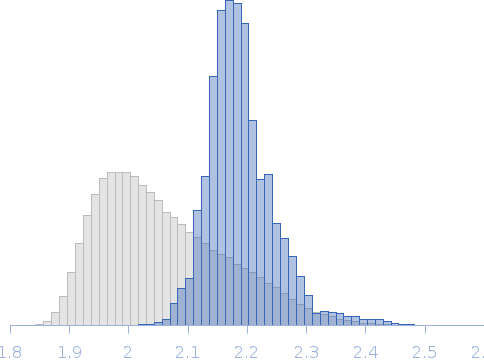 Rg, nm
Rg, nm