|
SAXS data (I(s) vs s, where s = 4πsinθ/λ and 2θ is the scattering angle) were collected on the SWING beam ine at the Soleil synchrotron (Gif-sur-Yvette, France). Samples were stored at 288 K in a robotic sample chamber and automatically loaded (22.3 mg/ml) onto a Superdex 200 Increase 3.2/300 gel-filtration column (GE Healthcare) equilibrated with 50 mM Tris HCl pH 7.5, 100 mM NaCl, 100 mM KCl, 10 mM MgCl2, 10 mM DTT and 5 % glycerol (v/v), at a flow rate of 150 μl min-1 by a HPLC (High performance liquid chromatography) instrument (Agilent), directly before elution into the sample detection chamber, where a monochromatic beam illuminated the sample with a wavelength of 0.1022 nm as it flowed through. The sample-detector distance was 1.79 m. During the elution, 250 scattering measurements were taken with 1.5-s time-frames and 0.5-s dead-time between frames. The in-house program FOXTROT (David and Pérez, 2009) was used to normalize and radially average the data. After averaging 20-30 buffer frames in PRIMUS (Konarev et al., 2003), the program DATASW (Shkumatov and Strelkov, 2015) was employed to (i) subtract the buffer average from each sample frame and (ii) calculation of I(0), Rg and MW. Frames corresponding to the peak where Rg was stable were used for averaging.
Concentration min = UNKNOWN
|
|
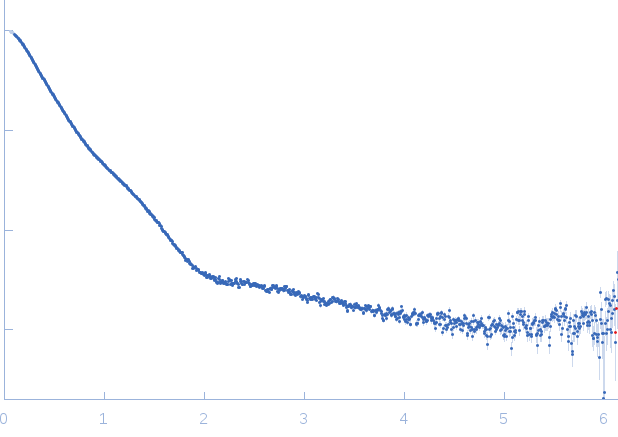 s, nm-1
s, nm-1
