| MWexperimental | 159 | kDa |
| MWexpected | 153 | kDa |
| VPorod | 218 | nm3 |
|
log I(s)
1.92×10-1
1.92×10-2
1.92×10-3
1.92×10-4
|
 s, nm-1
s, nm-1
|
|
|
|
|
|

|
|
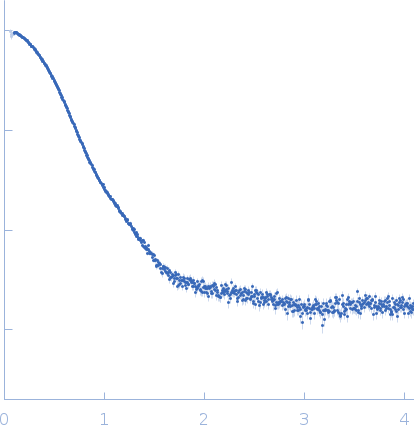
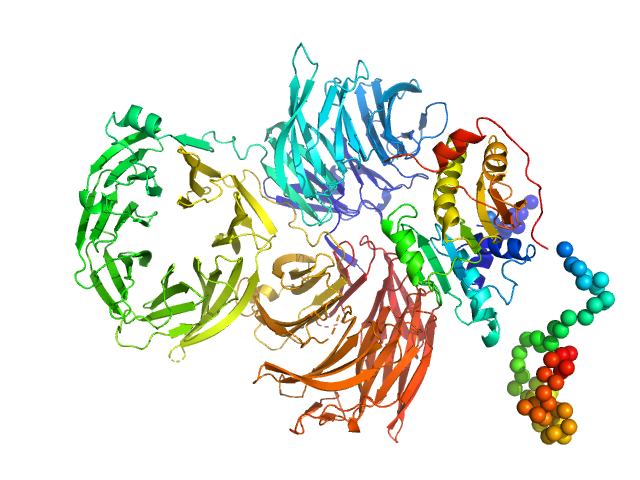
Synchrotron SAXS data from solutions of Yeast tRNA Nm34 methyltransferase Trm7-Trm734 complex in 50 mM HEPES-KOH, 200 mM, KCl, 5 % v/v glycerol, 10 mM β-mercaptoethanol, pH 8.0 were collected on the beam line BL-10C at the Phton Factory (PF; Tsukuba, Ibaraki, Japan) using a Pilatus3 2M detector at a sample-detector distance of 2.05 m and at a wavelength of λ = 0.1 nm (I(s) vs s, where s = 4πsinθ/λ and 2θ is the scattering angle). 40 images of 20 second frames were collected at 20°C. The data were normalized to the intensity of the incident beam and circularly averaged; the scattering of the solvent-blank was subtracted. The extrapolated scattering profile at a concentration of zero was calculated PRIMUS QT of ATSAS.
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||