|
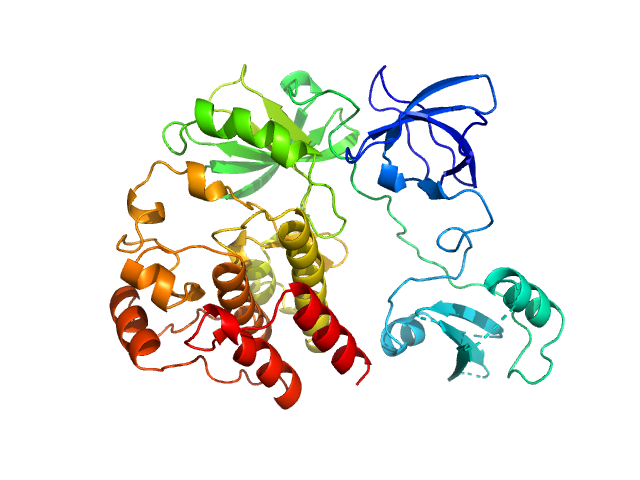
Synchrotron SAXS data for auto-inhibited module SH3-SH2-kinase domain (amino acids 214-659) wild-type of human Bruton’s tyrosine kinase (Btk) in 20 mM Tris-HCl, 150 mM NaCl, 5% Glycerol, 1 mM TCEP, pH 7.5 were collected on the BM29 beam line at the ESRF (Grenoble, France) using a Dectris Pilatus 1M detector at a sample-detector distance of 2.849 m and at a wavelength of λ = 0.099 nm (l(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). Solute concentrations ranging between 0.78 and 4.32 mg/ml were measured at 10°C. 10 successive 0.5 second frames were collected. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted. The raw curve measured from highest concentration was used for further analysis. The final scattering curve was analysed using PRIMUS software (ATSAS). The model depicts the DAMMIN ab initio reconstruction superimposed with the crystal structure 4XI2 (PDB). Ensemble optimisation method (EOM) was performed using three domains structure (1QLY, 2GE9 and 1K2P, PDB) to assess construct’s flexibility.
|
|
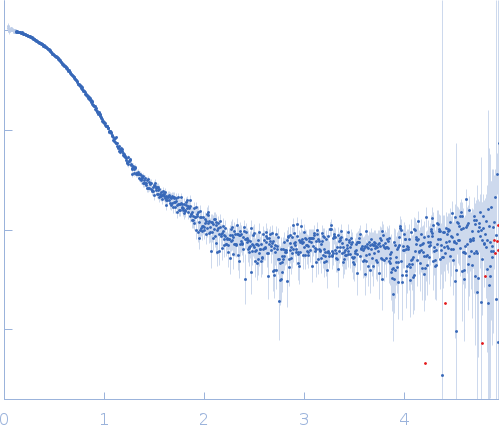 s, nm-1
s, nm-1
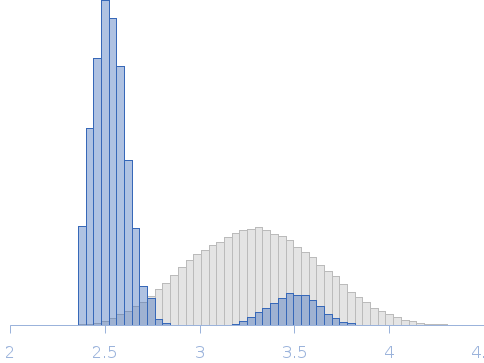 Rg, nm
Rg, nm