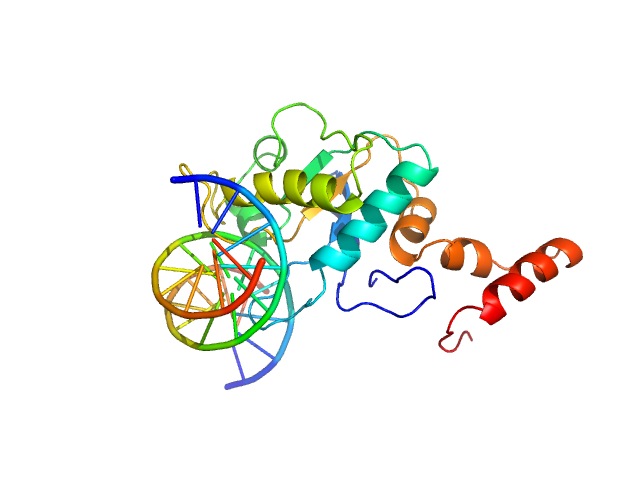|
Synchrotron SAXS data from solutions of EcoKMcrA restriction endonuclease (N-terminal domain) bound to 12bp oligoduplex DNA in 20 mM Tris–HCl pH 7.5, 200 mM KCl, 0.1 mM EDTA, 0.01% (w/v) sodium azide and 1 mM DTT, were collected on the EMBL P12 beam line at PETRA III (DESY, Hamburg, Germany) using a Pilatus 2M detector at a sample-detector distance of 3.1 m and at a wavelength of λ = 0.124 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). One solute concentration of 1.14 mg/ml was measured at 20°C. 20 successive 0.050 second frames were collected. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
Slightly prone to aggregation. NOTE! The DNA used in the model is homologous, and does not correspond in sequence, to the deposited DNA sequence of the complex analysed using SAXS.
|
|
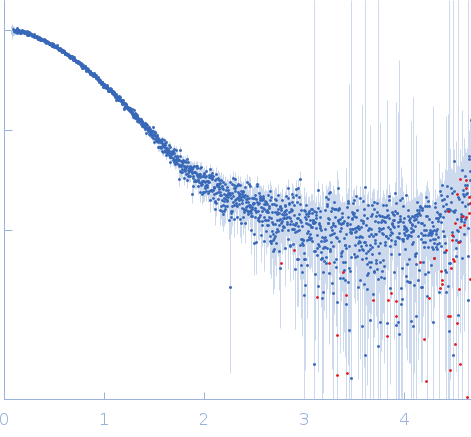 s, nm-1
s, nm-1