|
Synchrotron SAXS
data from solutions of
Matrix protein from Newcastle disease virus at neutral pH
in
STE buffer 100 mM NaCl, 10 mM Tris-HCl, and 1 mM EDTA, pH 4
were collected
on the
EMBL P12 beam line
at the PETRA III storage ring
(DESY; Hamburg, Germany)
using a Pilatus 2M detector
at a sample-detector distance of 3 m and
at a wavelength of λ = 0.124 nm
(I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle).
One solute concentration of 2.20 mg/ml was measured
at 20°C.
20 successive
0.045 second frames were collected.
The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
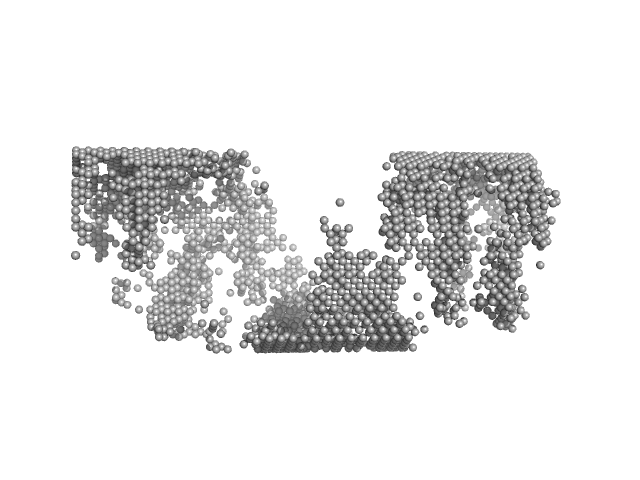
The scattering from dimers was subtracted from the curve used in Dammin to focus only on large molecular weight species (helices).
|
|
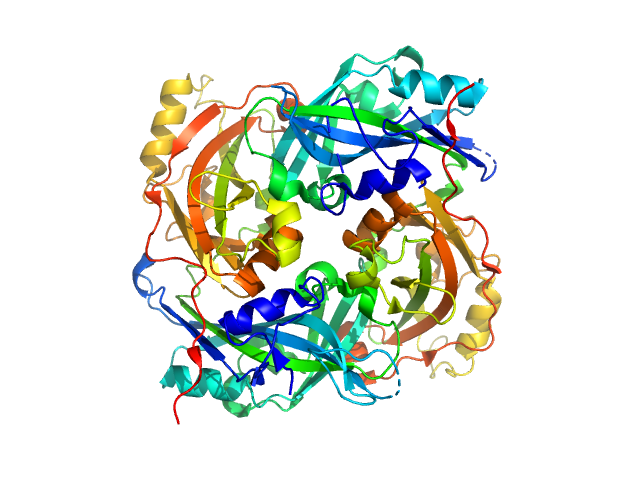
Matrix protein
|
| Mol. type |
|
Protein |
| Organism |
|
Newcastle disease virus (strain Chicken/Australia-Victoria/32) |
| Olig. state |
|
Unknown |
| Mon. MW |
|
39.7 kDa |
| |
| UniProt |
|
P11206
|
| Sequence |
|
FASTA |
| |
|
PDB ID
|
|
4G1L
|
| |
|
 s, nm-1
s, nm-1