|
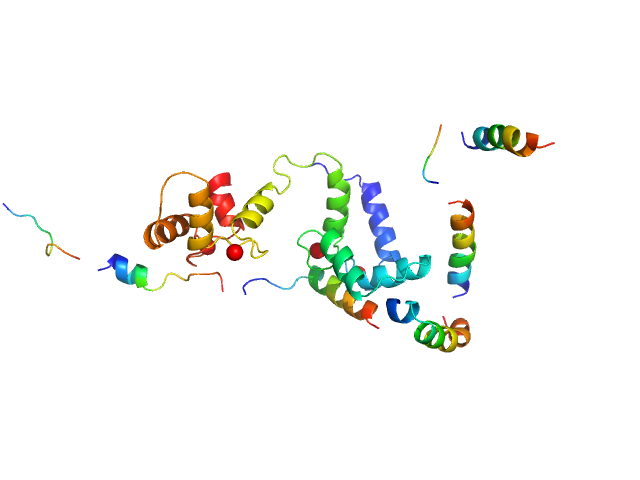
SAXS data from a 1:1 complex formed between a 73% deuterated 4Ca2+-CaM and the HIV-1 MA protein in 100% D2O, 50 mM MOPS pH 7.0, 2 mM CaCl2 with 2 mM TCEP were collected using a Bruker Nanostar instrument at the Bragg Institute (Australian Nuclear Science and Technology Organisation, Lucas Heights, Australia) equipped with a VANOTEC-2000 detector (I(s) vs s, where s = 4π sin θ/λ and 2θ is the scattering angle; λ=0.15406 nm). Approximately 15 µL of a 7.2 mg/ml protein solution was loaded into a quartz capillary mounted in a stainless steel holder. Sample and buffer were acquired for equal times. The data were radially averaged, and the scattering of the solvent-blank was subtracted. The SAXS data were placed on an absolute scale using the scattering from water as the standard. Small-angle neutron scattering (SANS) data with contrast variation were also collected on the QUOKKA instrument also at Bragg Institute, yielding a 4-point contrast series. The combined SAXS and SANS data were used with the program MONSA to produce an ab intio shape model depicting the relative dispositions of the two proteins as well as a rigid body atomistic model (refer to NOTES, below). These models are shown here, together with the X-ray data and corresponding fits. SANS data with details of the data collection, analysis, modelling protocols and model fits are contained in a downloadable zip archive.
NOTES: The rigid-body atomistic representation/model as displayed here included the full-length Ca2+-CaM and the MA sequence spanning amino acids 1-113, i.e. does not include the flexible C-terminal tail (amino acids 114-133).
|
|
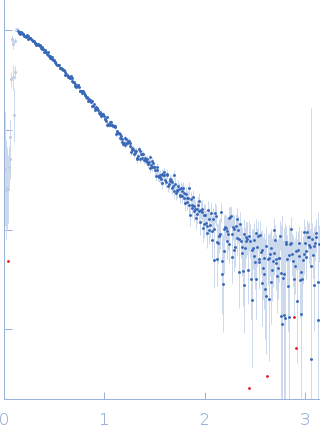 s, nm-1
s, nm-1