|
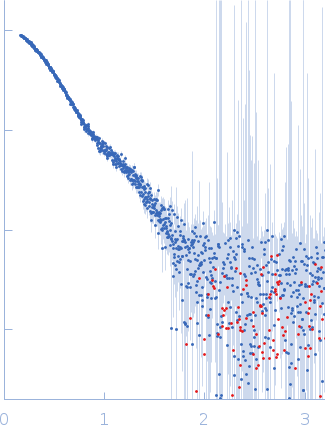
Synchrotron SAXS data from solutions of riboflavin biosynthesis protein (RibD) in 150 mM NaCl, 10 mM Tris, 1 mM DTT, 5% v/v glycerol, pH 7.4 were collected on the EMBL P12 beam line at PETRA III (DESY; Hamburg, Germany) using a Pilatus 6M detector at a sample-detector distance of 3 m and at a wavelength of λ = 0.124 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). One solute concentration of 1.60 mg/ml was measured at 20°C. 20 successive 0.100 second frames were collected. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
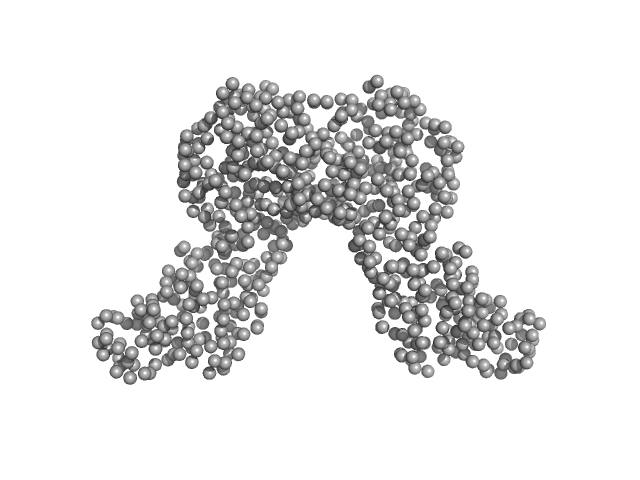
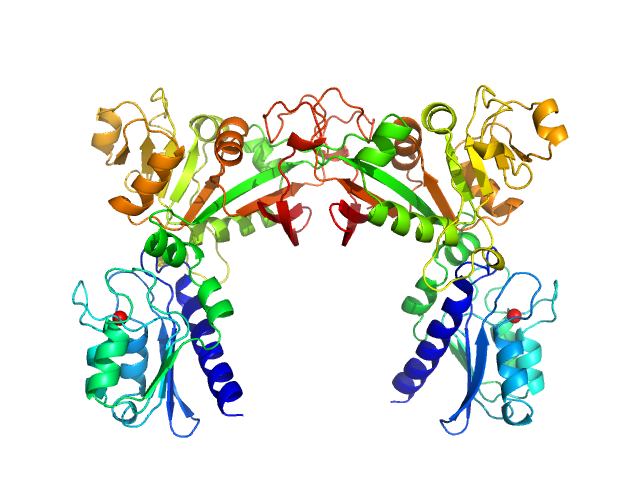
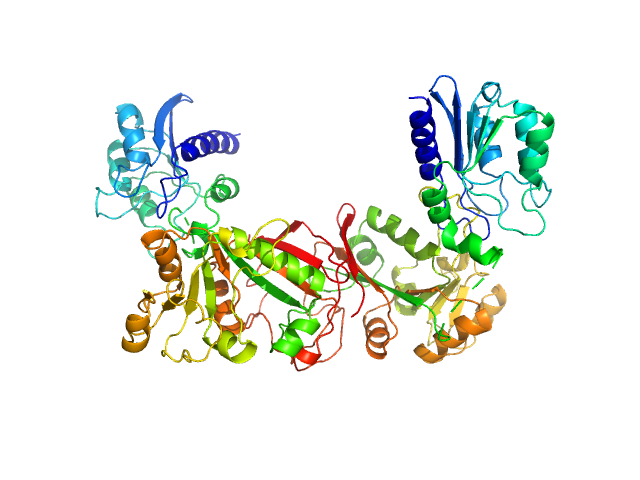
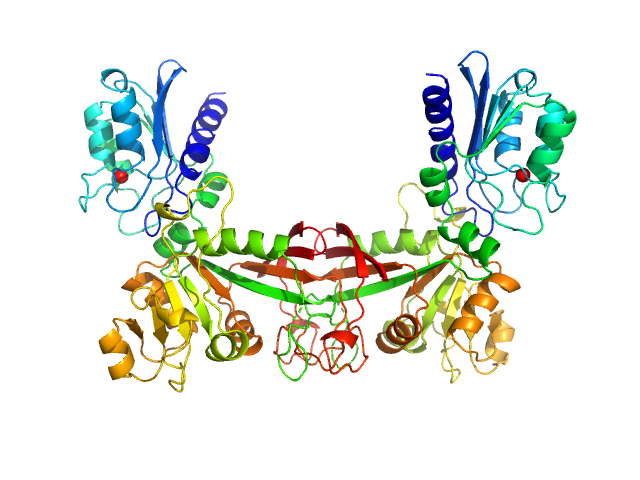
Data have been truncated at low-s to remove the effects on the scattering caused by slight non-specific aggregation. From the data displayed in this entry, the concentration independent MW estimates fall in the range of 75–87 kDa. The models displayed in this entry are: Top - a single GASBOR model representative; Second from top - a SASREF refined rigid-body model in P2 symmetry; Third - a SREFLEX normal mode refined model in P1 symmetry (allowing for non-symmetry related movements in the peripheral zinc-binding domain lobes) and; Fourth - the fit to the SAXS data of a closely related Acinetobacter baumannii sp. RibD dimer (PDB, 3ZPG; MW = 77 kDa; UniProt: D0CB74; 97% sequence identity). Additional models and fits, and the full SAXS profile are made available in the full entry zip archive.
|
|
 s, nm-1
s, nm-1