|
Synchrotron SAXS data from solutions of fibronectin 2 and 3 of cell adhesion molecule L1 (L1CAM; expressed in expressed in HEK293F cells with kifunensine) in 50 mM MES pH 6.5, 150 mM NaCl, 5% glycerol, were collected on the EMBL P12 beam line at the PETRA III storage ring (Hamburg, Germany) using a Pilatus 6M detector at a sample-detector distance of 3 m and at a wavelength of λ = 0.124 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). In-line size-exclusion chromatography (SEC) SAS was employed. The SEC parameters were as follows: A 40.00 μl sample at 10 mg/ml was injected at a 0.35 ml/min flow rate onto a GE Superdex 200 Increase 5/150 column at 20°C. 2880 successive 0.250 second frames were collected through the column elution. 81 SAXS data frames from the top of a single SEC elution peak were selected for averaging. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
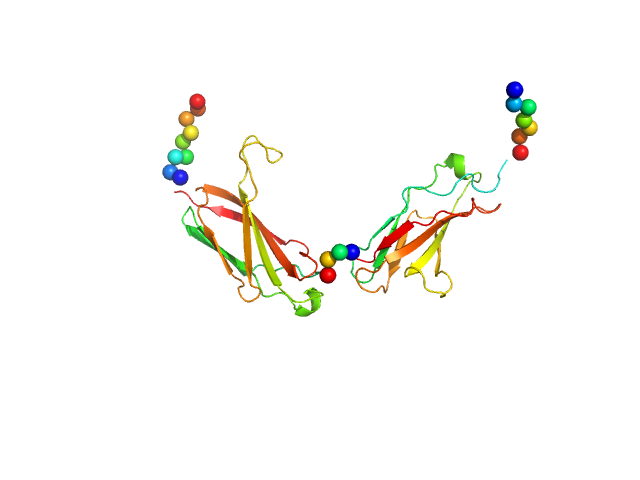
The entry shows 1) the individual ab initio reconstruction, 2) the spatial reconstruction of the averaged model and 3) the rigid body model with the best fitting showing the two fibronectin domains in a bent conformation that differs to the elongated conformation seen in the crystal structure (PDB entry 8AFO or 8AFP). SEC-SAXS data were collected with the same parameters from three samples differing mainly in the glycosylation pattern (refer to SASBDB entries SASDPB6 and SASDPC6).
|
|
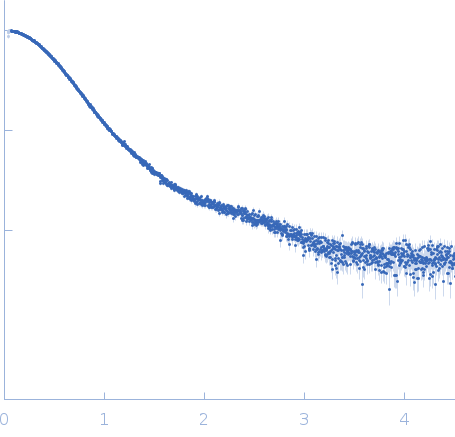 s, nm-1
s, nm-1