|
Synchrotron SAXS data from solutions of the receptor-type tyrosine-protein phosphatase mu extracellular domains (PTPRM-ECDs) in 50 mM MES, 250 mM NaCl, 3% v/v glycerol, pH 6 were collected on the EMBL P12 beam line at PETRA III (DESY, Hamburg, Germany) using a Pilatus 6M detector at a sample-detector distance of 3 m and at a wavelength of λ = 0.124 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). In-line size-exclusion chromatography (SEC) SAS was employed. The SEC parameters were as follows: A 40.00 μl sample at 2.3 mg/ml was injected at a 0.35 ml/min flow rate onto a GE Superdex 200 Increase 5/150 column at 20°C. 2880 successive 0.250 second frames were collected through the entire column elution; 54 data frames from the main sample peak were used for averaging. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted (processed using CHROMIXS).
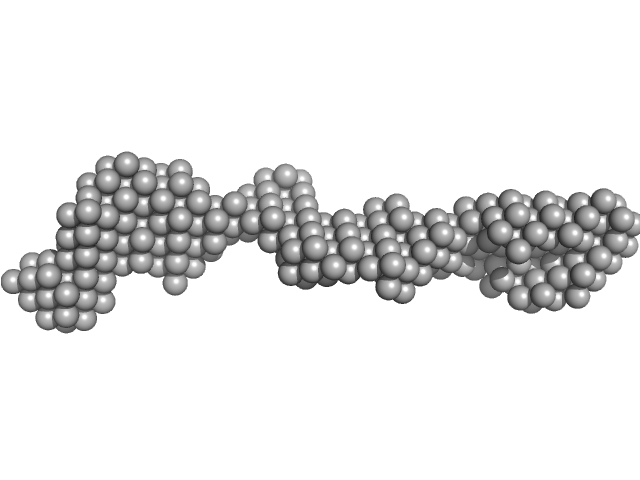
The quoted experimental molecular weight of 94 kDa was obtained using a Bayesian inference concentration independent method from the SAXS data. The corresponding experimental MALLS molecular weight estimate is 100 kDa, that includes a conjugate analysis of the protein molecular weight of (80 kDa) and glycan component (19 kDa). The corresponding SEC-SAXS and SEC-MALLS-QELS data as well as the results from DAMMIN ab initio, and hybrid CORAL/Alphafold/Xray crystal structure rigid-body modelling are made available in the full entry zip archive.
|
|
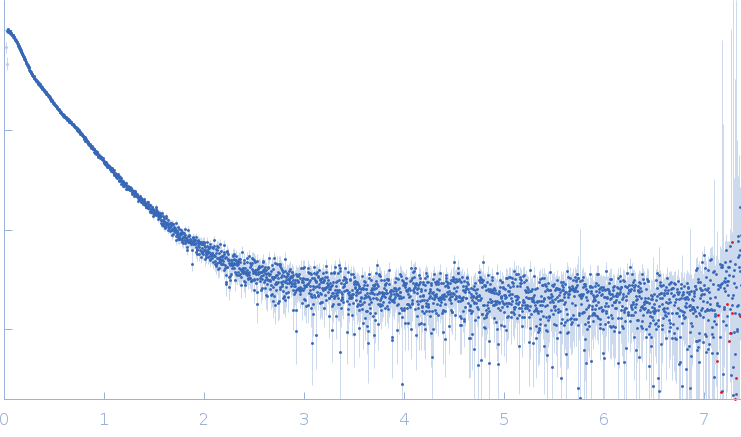 s, nm-1
s, nm-1