| MWexperimental | 173 | kDa |
| MWexpected | 173 | kDa |
| VPorod | 243 | nm3 |
|
log I(s)
1.05×100
1.05×10-1
1.05×10-2
1.05×10-3
|
 s, nm-1
s, nm-1
|
|
|
|

|
|
|
|
|
|
|
|
|
|
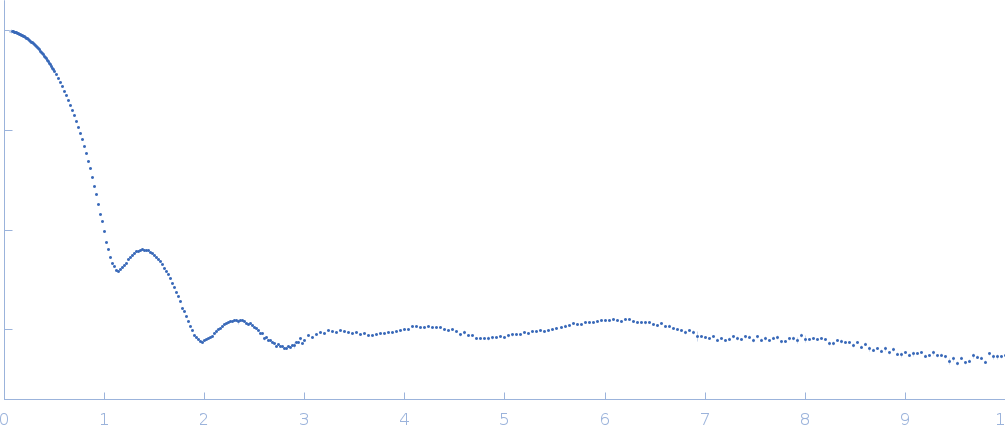
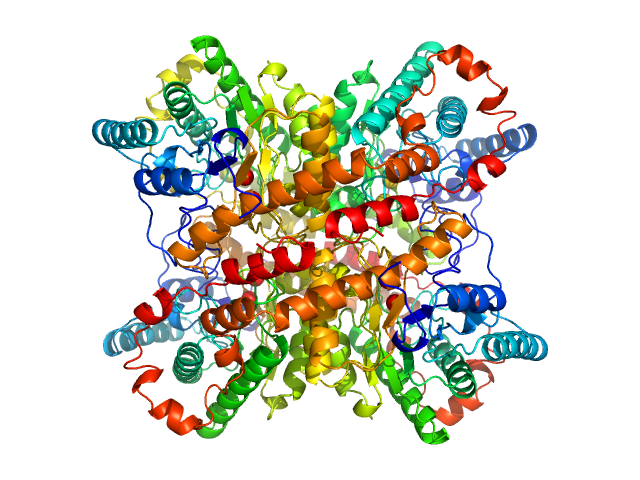
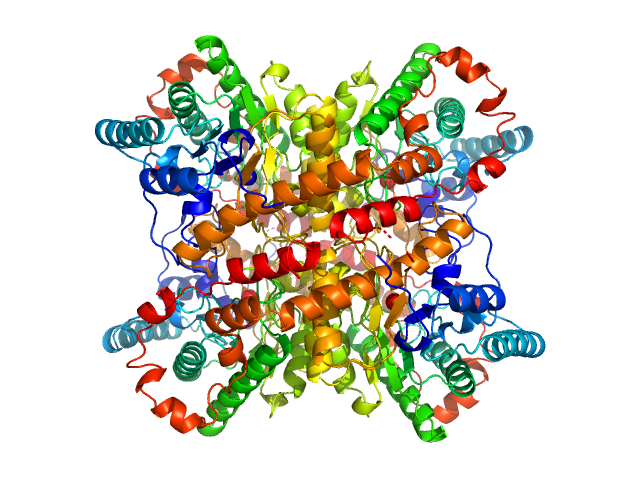
The consensus SAXS profile for xylose isomerase was generated using the datcombine tool (ATSAS 3.1.0) with outlier-filter applied. The data input to datcombine were fourteen scattering profiles made up of pure SEC-SAXS (5), pure batch SAXS (6) and merged SEC-SAXS-batch SAXS (3) data. All contributing data were independent measurements, and no individual measurement was represented more than once in the contributing scattering profile set. The buffer for substantial majority of the contributing data was 50 mM Tris, pH 7.5, 100 mM NaCl, 1 mM MgCl2. Protein concentrations for batch measurements ranged from 0.5 - 21.5 mg/mL, and all batch data > 1 mg/mL were merged with either SEC-SAXS or lower concentration data demonstrated to be free of interparticle interference, which is known to be significant for xylose isomerase concentrations > 1.0 mg/mL. The xylose isomerase atomistic model for CRYSOL, Pepsi-SAXS and FoXS calculations was derived from PDB ID 1MNZ tetramer with an N-terminal Met added using PyMol, and small-molecule crystallisation agents removed. Custom WAXSiS calculations (with Gromacs software) used the same coordinates and added explicit waters and appropriate number of ions for the MD calculation.
The data input to datcombine are made available for download in the associated zip file. Model fits are shown in order (top to bottom): DAMMIN, CRYSOL, Pepsi-SAXS, FoXS, and custom WAXSiS. The unusually good statistics for the consensus SAXS data generally give rise to large χ-square values for the model fits.
Tags:
benchmark
|
|
|||||||||||||||||||||||||||||||||||||||