Structural and functional insights into peptidoglycan access for the lytic amidase LytA of Streptococcus pneumoniae.
Mellroth P,
Sandalova T,
Kikhney A,
Vilaplana F,
Hesek D,
Lee M,
Mobashery S,
Normark S,
Svergun D,
Henriques-Normark B,
Achour A
MBio
5(1):e01120-13
(2014 Feb 11)
|
|
|
|
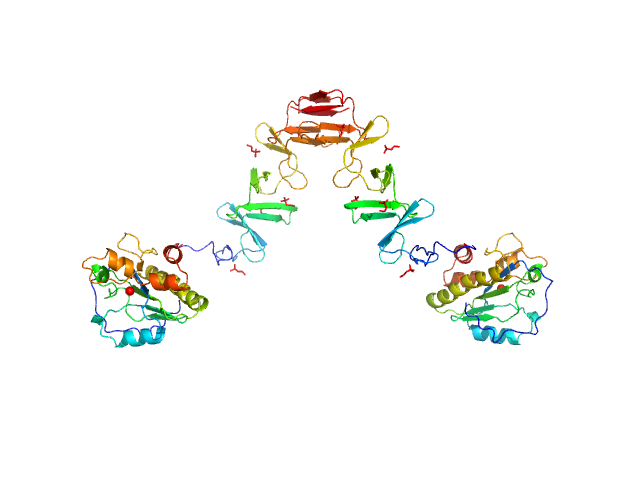
|
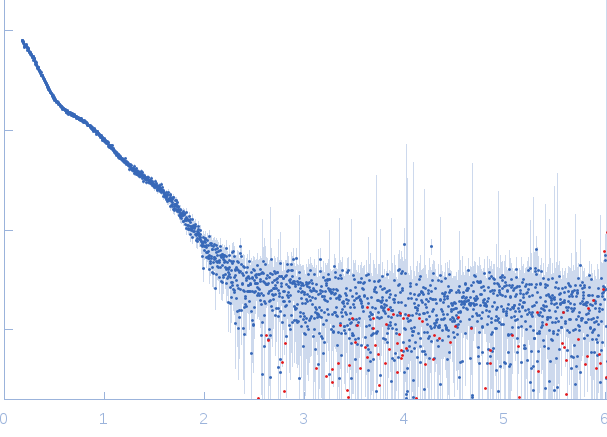
| Sample: |
Lytic Amidase with choline dimer, 75 kDa Streptococcus pneumoniae protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 5 mM choline chloride 1 µM ZnCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Jul 2
|
|
| RgGuinier |
4.9 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
115 |
nm3 |
|
|
|
|
|
|
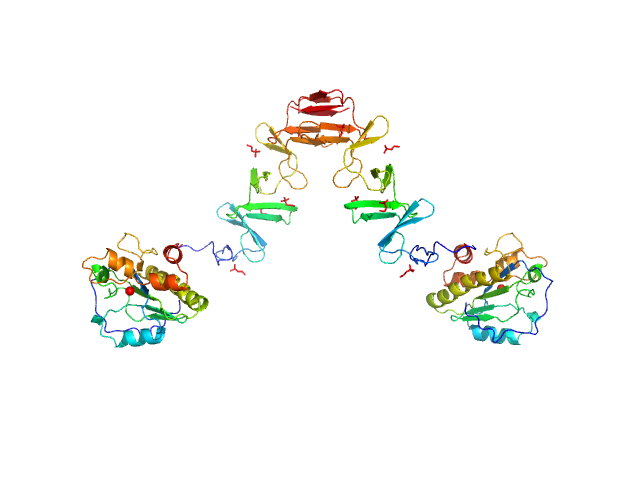
|
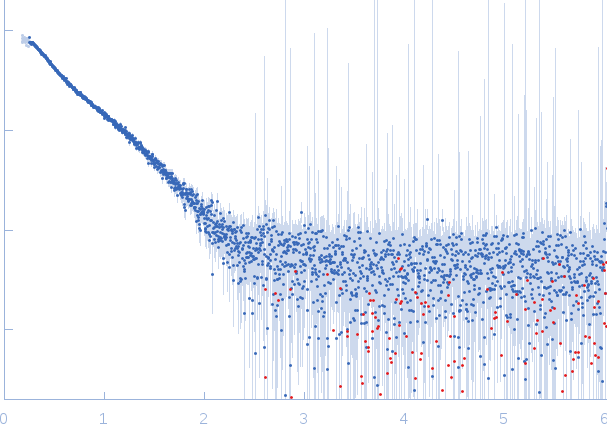
| Sample: |
Prophage Lytic Amidase with choline dimer, 73 kDa Streptococcus pneumoniae protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 5 mM choline chloride 1 µM ZnCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Jul 2
|
|
| RgGuinier |
5.9 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
130 |
nm3 |
|
|
|
|
|
|
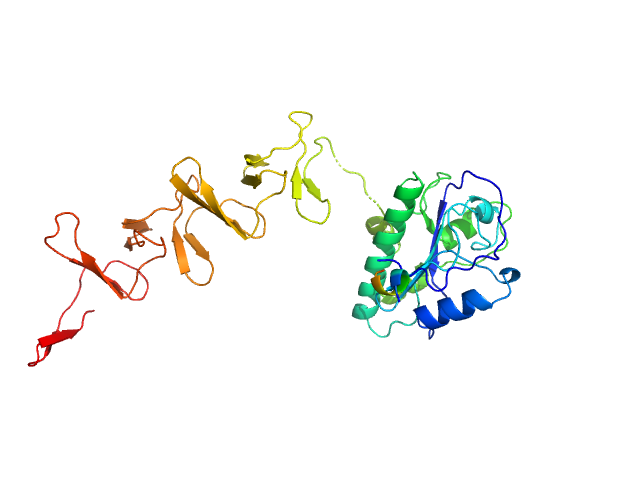
|
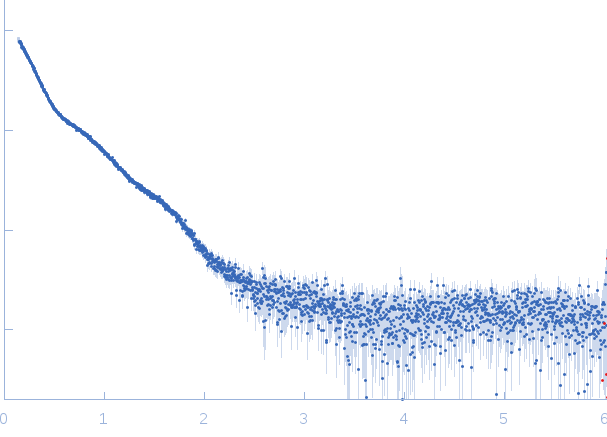
| Sample: |
Lytic Amidase without choline monomer, 37 kDa Streptococcus pneumoniae protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 1 µM ZnCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Jul 3
|
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
|
|
|
|
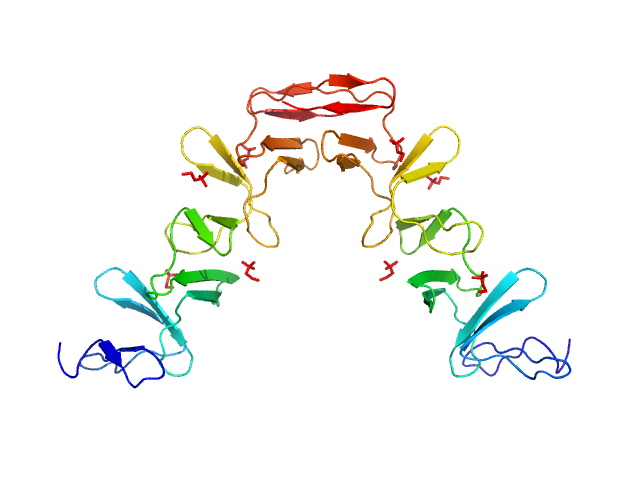
|
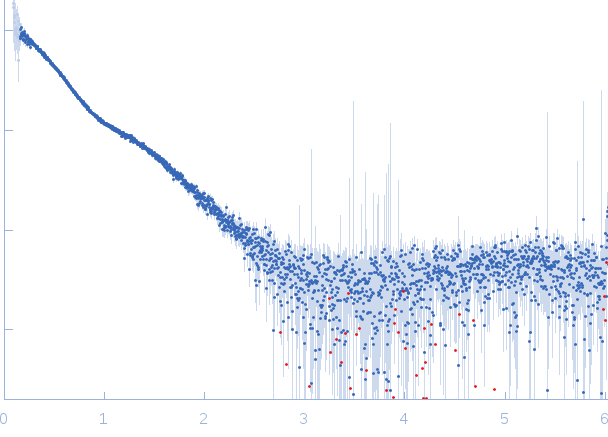
| Sample: |
Lytic Amidase choline-binding domain dimer, 17 kDa Streptococcus pneumoniae protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 5 mM choline chloride 1 µM ZnCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Dec 11
|
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
49 |
nm3 |
|
|