Stability of local secondary structure determines selectivity of viral RNA chaperones.
Bravo JPK,
Borodavka A,
Barth A,
Calabrese AN,
Mojzes P,
Cockburn JJB,
Lamb DC,
Tuma R
Nucleic Acids Res
(2018 May 18)
|
|
|
|
|
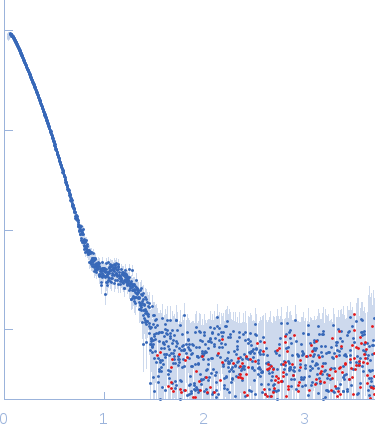
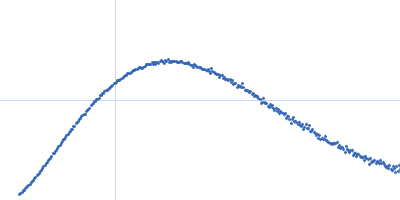
| Sample: |
Nonstructural protein sigma NS octamer, 325 kDa Avian orthoreovirus protein
20mer RNA (unstructured) dimer, 13 kDa RNA
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Feb 25
|
|
| RgGuinier |
7.8 |
nm |
| Dmax |
38.0 |
nm |
| VolumePorod |
964 |
nm3 |
|
|
|
|
|
|
|
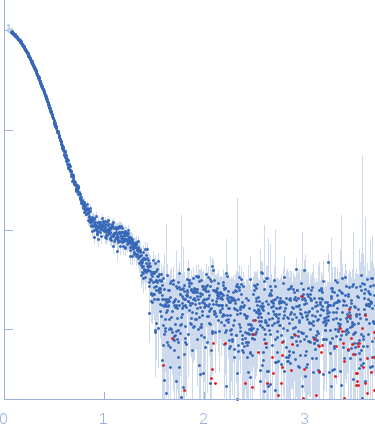
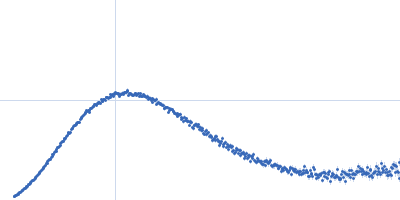
| Sample: |
Nonstructural protein sigma NS hexamer, 244 kDa Avian orthoreovirus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Feb 25
|
|
| RgGuinier |
5.5 |
nm |
| Dmax |
23.1 |
nm |
| VolumePorod |
670 |
nm3 |
|
|