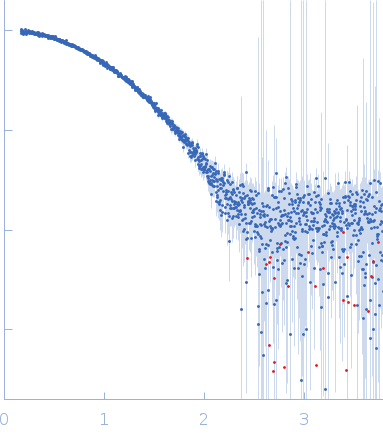
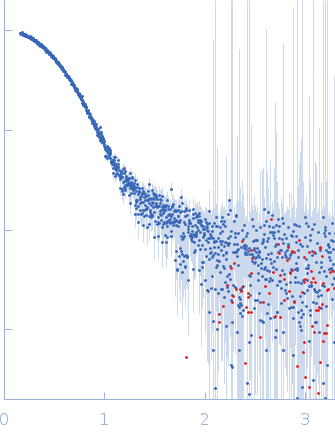
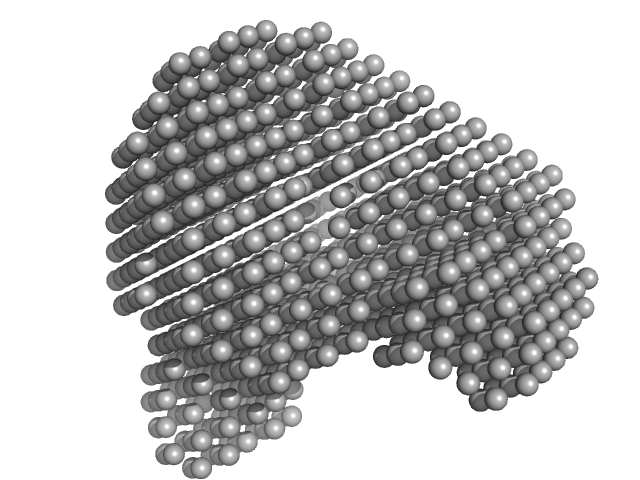
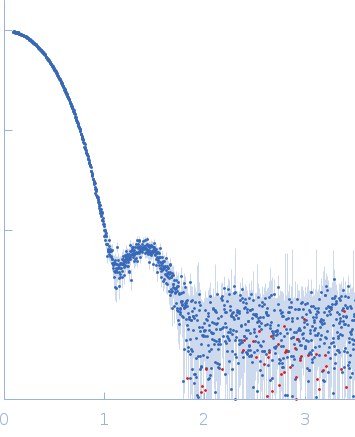
Correlation Map, a goodness-of-fit test for one-dimensional X-ray scattering spectra.
Franke D, Jeffries CM, Svergun DINat Methods 12(5):419-22 (2015 May)
|
PMID: 25849637
doi: 10.1038/nmeth.3358 |
Published in SASBDB: |
|
|||||||||||||||||||||||
|
|||||||||||||||||||||||
|
|||||||||||||||||||||||