|
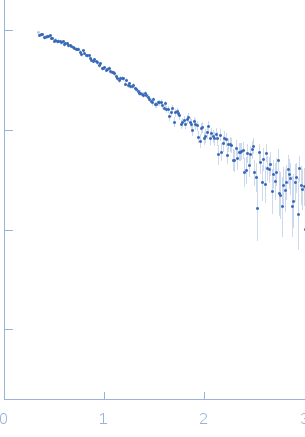
SAXS data from solutions of telomeric DNA duplex in 20 mM Tris-HCl, 50 mM LiCl, pH 7.5 were collected using a Rigaku BioSAXS-1000 instrument at CEITEC (Brno, Czech Republic) equipped with a Pilatus 100K detector at a sample-detector distance of 0.5 m (I(s) vs s, where = 4πsinθ/λ; 2θ is the scattering angle and λ = 0.154 nm). Six successive 3600 second frames were collected at a sample temperature of 4°C and a solute concentration of 1.4 mg/ml. The data were normalized to the intensity of the transmitted beam and radially averaged and the corresponding scattering from the solvent-blank was subtracted to produced the scattering profile displayed in this entry.
Refer to the related SASBDB entries SASDB79 and SASDC62.
|
|
 s, nm-1
s, nm-1
