|
|
|
|
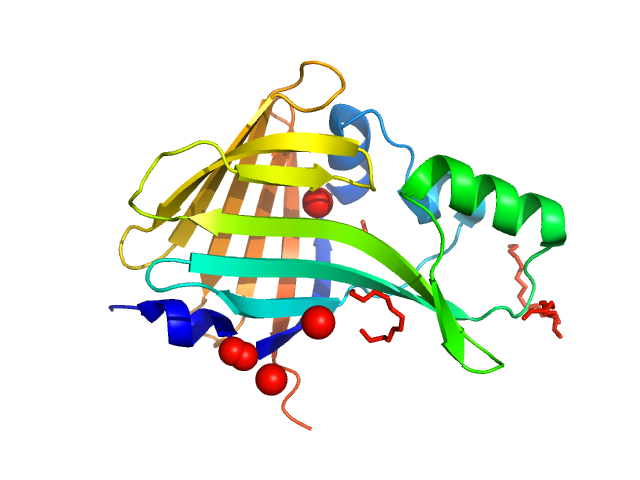
|
| Sample: |
Oplophorus-luciferin 2-monooxygenase catalytic subunit monomer, 20 kDa Oplophorus gracilirostris protein
|
| Buffer: |
10 mM Tris-HCl, 50 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, CEITEC on 2021 May 27
|
Illuminating the mechanism and allosteric behavior of NanoLuc luciferase.
Nat Commun 14(1):7864 (2023)
Nemergut M, Pluskal D, Horackova J, Sustrova T, Tulis J, Barta T, Baatallah R, Gagnot G, Novakova V, Majerova M, Sedlackova K, Marques SM, Toul M, Damborsky J, Prokop Z, Bednar D, Janin YL, Marek M
|
| RgGuinier |
1.8 |
nm |
| Dmax |
5.9 |
nm |
| VolumePorod |
38 |
nm3 |
|