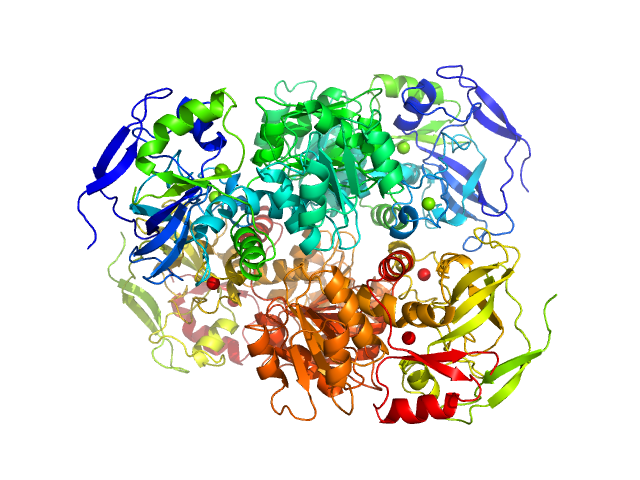
|
Synchrotron SAXS data from alcohol dehydrogenase 1 in 50 mM HEPES, 150 mM NaCl, 2% v/v glycerol, pH 7 were collected on the EMBL P12 beam line at PETRA III (DESY, Hamburg, Germany) using a Pilatus 6M detector at a sample-detector distance of 3 m and at a wavelength of λ = 0.124 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). In-line size-exclusion chromatography (SEC) SAS was employed. The SEC parameters were as follows: A 100.00 μl sample at 9.2 mg/ml was injected at a 0.50 ml/min flow rate onto a GE Superdex 200 Increase 10/300 column at 20°C. 67 successive 1 second frames were collected through the major SEC-elution peak and processed using CHROMIXS. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
Alcohol dehydrogenase 1 underwent pre-purification prior to SEC-SAXS using the following method. All procedures were performed at 4 oC. The protein (from Sigma; Gel Filtration Markers Kit MWGF1000) was made to approximately 25 mg/ml in 25 mM HEPES, 50 mM NaCl, 5 mM urea, 1% v/v glycerol, pH 7. Approximately 200 μl of sample were loaded onto a Superdex 75 Increase 10/300 column (GE Healthcare) equilibrated in the same buffer (flow rate = 0.4 ml/min). Fractionated aliquots corresponding to the highest absorbing peak (estimated using UV A280 and UV A245 nm) were pooled and concentrated (30 kDa centrifuge spin filter) to a final concentration of 9.2 mg/ml (the concentration was determined from triplicate UV A280 measurements using an E0.1% of 1.326 (= 1 g/l) calculated from the amino acid sequence (ProtParam)). Approximately 50 μl aliquots were snap-frozen in liquid nitrogen then stored at -80oC prior to the SEC-SAXS analysis that was performed at room temperature in 50 mM HEPES, 150 mM NaCl, 2% v/v glycerol, pH 7. The Rg-correlation through the SEC-SAXS peak, the individual unsubtracted SEC-SAXS frames as well as the results from coupled MALLS and QELS analysis are included in the full entry zip archive. The quoted experimental molecular weight was determined using MALLS in combination with refractive-index (RI) measurements that were recorded from the same sample eluting from the column using a split-flow SEC-SAXS-light scattering configuration (Graewert et al., (2015) Sci. Reports. 5, 10734: doi: 10.1038/srep10734). The average hydrodynamic radius of the protein is 4.5 nm.
|
|
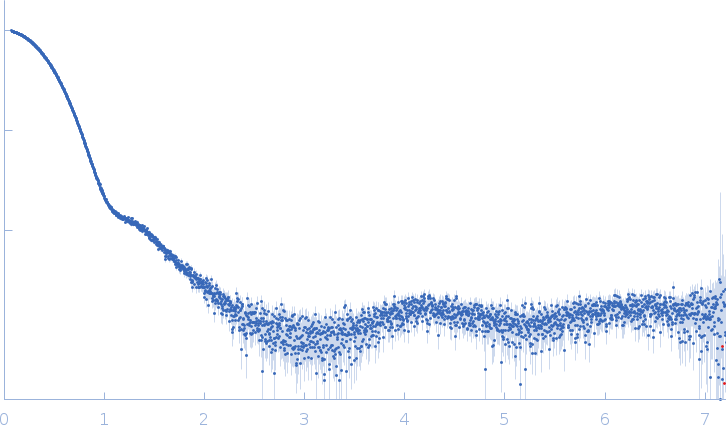 s, nm-1
s, nm-1