|
|
|
|
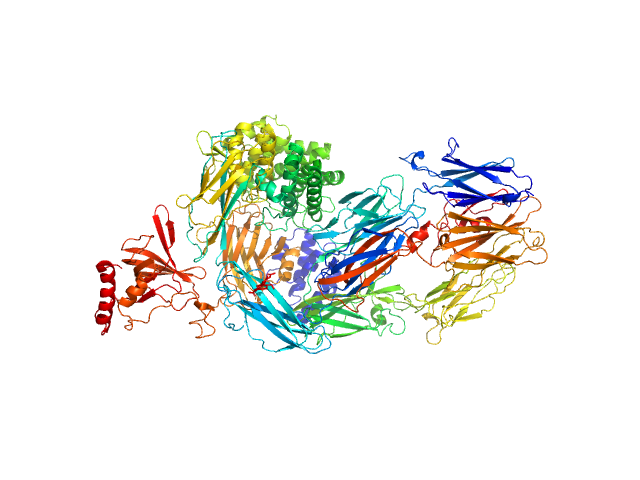
|
| Sample: |
Complement C5 monomer, 186 kDa Homo sapiens protein
|
| Buffer: |
20mM Tris pH, 75mM NaCl, and 3% glycerol, pH: 7.4
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Sep 12
|
The allosteric modulation of Complement C5 by knob domain peptides.
Elife 10 (2021)
...Graewert MA, Birtley JR, Schulze ME, Crennell S, Robinson SA, Holmes B, Oleinikovas V, Nilsson PH, Snowden J, Ellis V, Mollnes TE, Deane CM, Svergun D, Lawson AD, van den Elsen JM
|
| RgGuinier |
4.8 |
nm |
| Dmax |
17.6 |
nm |
| VolumePorod |
392 |
nm3 |
|