|
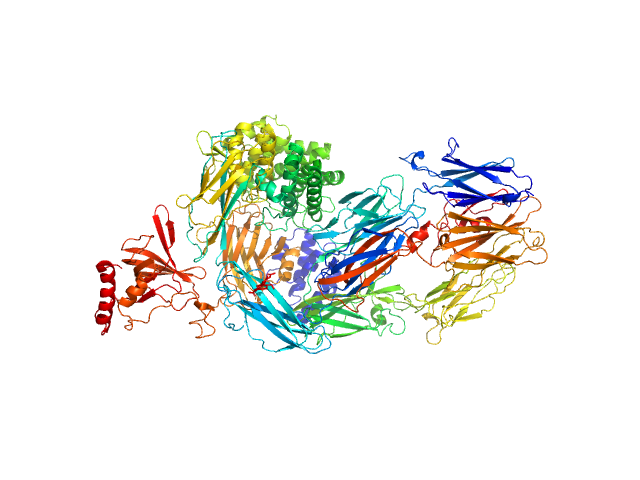
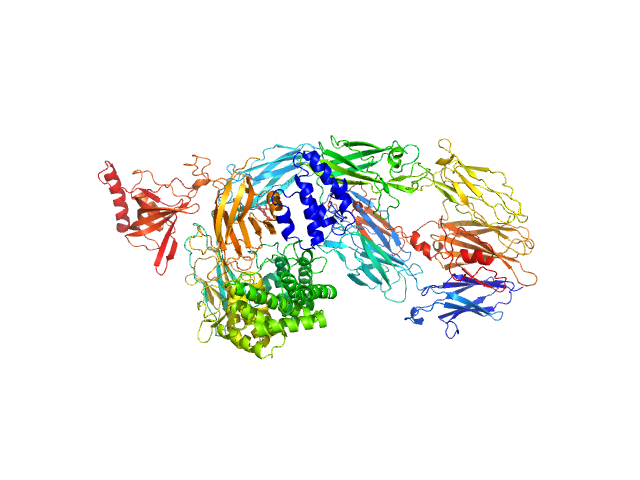
Data was collected at the EMBL P12 beam line (PETRA III, DESY Hamburg, Germany). Data was collected with inline in size exclusion chromatography (SEC) mode using the Agilent 1260 Infinity II Bio-inert LC system at the beamline. 50 μL of complement component C5 at 31.6 μM (5.96 mg/ml) were injected onto a Superdex 200 Increase 5/150 column (GE Healthcare) at a flow rate of 0.35 ml/min. The mobile phase was comprised of 20mM Tris pH 7.35, 75mM NaCl, and 3% glycerol. The column elute was directly streamed to the SAXS capillary cell, and throughout the 15-minute-run 900 frames of 1 sec exposure were collected. After data reduction and radial averaging the program CHROMIXS was employed. Around 100 statistically similar buffer frames were selected and used for background subtraction of the sample frames from the chromatographic peak. This results in the final I(s) vs s scattering profiles, where s = 4πsinθ/λ, 2θ is the scattering angle and λ = 1.24 Å. The scattering data in the momentum transfer range 0.05 < s < 0.32 nm-1 were collected with a PILATUS 6M pixel detector at a distance of 3.1 m from the sample.
On the same day, multi-angle laser light scattering (MALLS) data was collected with a separate SEC run yet under the same experimental conditions (set-up, buffer, run parameters etc.).A Wyatt Technologies miniDAWN TREOS multiangle laser light scattering detector was used, coupled to an OptiLab T-Rex differential refractometer for protein concentration determination (dn/dc was taken as 0.185). The MALLS system was calibrated relative to the scattering from toluene. In addition to the apo entry, ligated versions as described in the manuscript are available as additional files.
Storage temperature = UNKNOWN
|
|
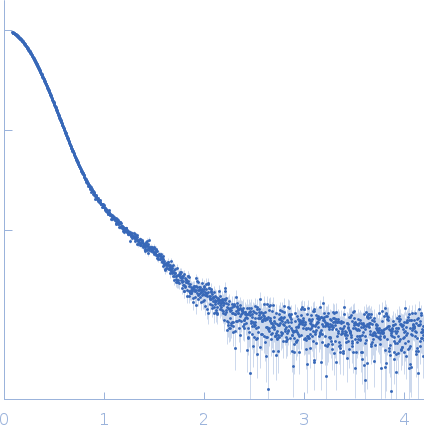 s, nm-1
s, nm-1