Cooperation between intrinsically disordered and ordered regions of Spt6 regulates nucleosome and Pol II CTD binding, and nucleosome assembly.
Kasiliauskaite A,
Kubicek K,
Klumpler T,
Zanova M,
Zapletal D,
Koutna E,
Novacek J,
Stefl R
Nucleic Acids Res
(2022 May 30)
|
|
|
|
|
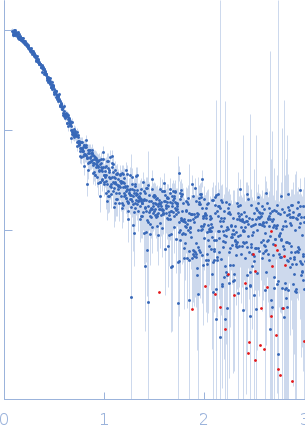
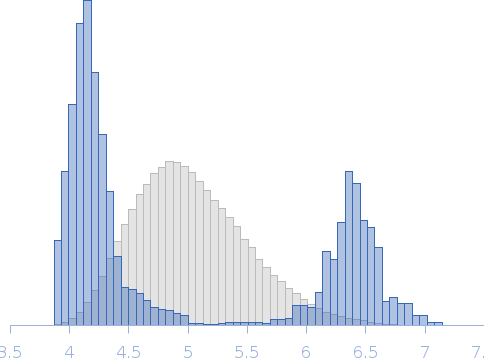
| Sample: |
Transcription elongation factor SPT6 monomer, 145 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
25 mM NaPi; 150mM NaCl; 0.5 mM EDTA; 5% glycerol; 1 mM DTT, pH: 7.5 |
| Experiment: |
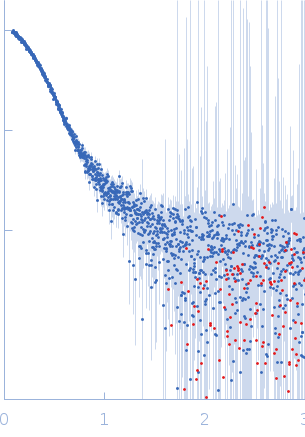
SAXS
data collected at EMBL P12, PETRA III on 2015 Oct 22
|
|
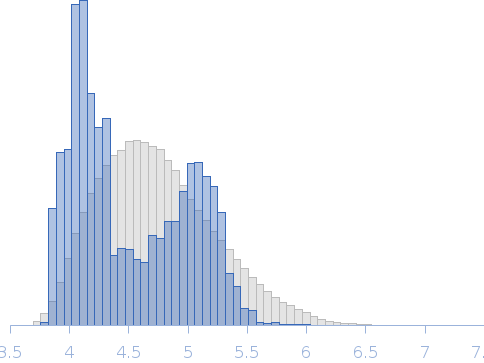
| RgGuinier |
4.5 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
249 |
nm3 |
|
|
|
|
|
|
|
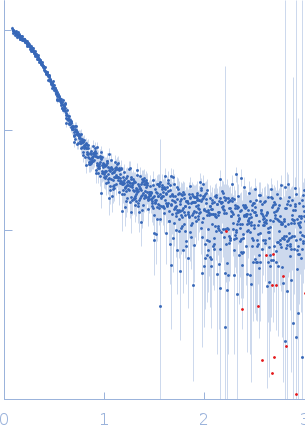
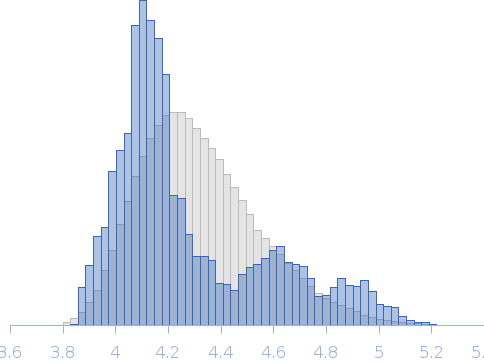
| Sample: |
Transcription elongation factor SPT6 - ΔtSH2 variant monomer, 122 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
25 mM NaPi; 150mM NaCl; 0.5 mM EDTA; 5% glycerol; 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Oct 22
|
|
| RgGuinier |
4.1 |
nm |
| Dmax |
13.9 |
nm |
| VolumePorod |
245 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Transcription elongation factor SPT6 - ΔN Spt6 variant monomer, 132 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
25 mM Hepes; 150 NaCl; 0.5 mM EDTA; 5% glycerol; 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Oct 3
|
|
| RgGuinier |
4.7 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
288 |
nm3 |
|
|