Npl3 functions in mRNP assembly by recruitment of mRNP components to the transcription site and their transfer onto the mRNA.
Keil P,
Wulf A,
Kachariya N,
Reuscher S,
Hühn K,
Silbern I,
Altmüller J,
Keller M,
Stehle R,
Zarnack K,
Sattler M,
Urlaub H,
Sträßer K
Nucleic Acids Res
(2022 Dec 30)
|
|
|
|
|
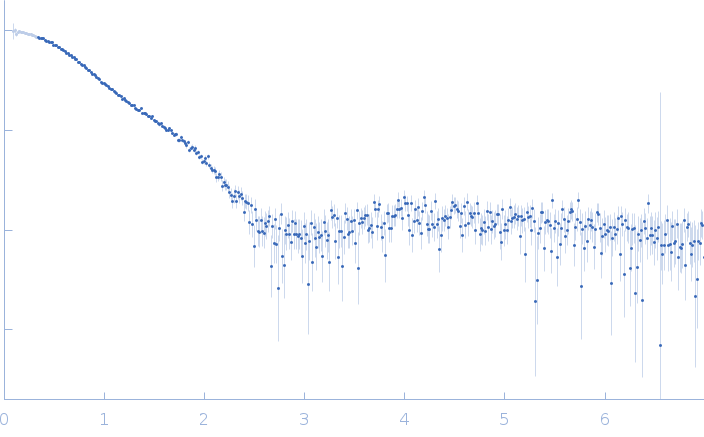
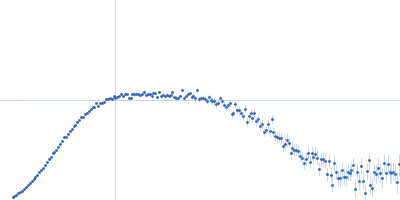
| Sample: |
Serine/arginine (SR)-type shuttling mRNA binding protein NPL3 monomer, 18 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
20 mM NaPO4, 50 mM NaCl, 1 mM DTT, pH: 6.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, SFB 1035, Technische Universität München on 2020 Feb 20
|
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.8 |
nm |
| VolumePorod |
24 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Serine/arginine (SR)-type shuttling mRNA binding protein NPL3 monomer, 18 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
20 mM NaPO4, 50 mM NaCl, 1 mM DTT, pH: 6.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, SFB 1035, Technische Universität München on 2020 Feb 20
|
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
25 |
nm3 |
|
|
|
|
|
|
|
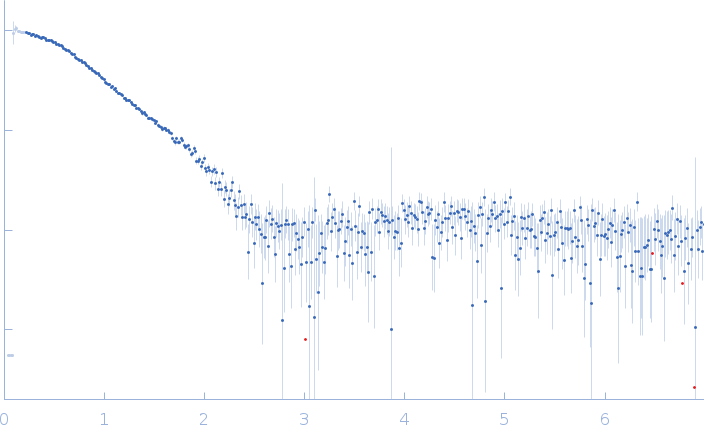
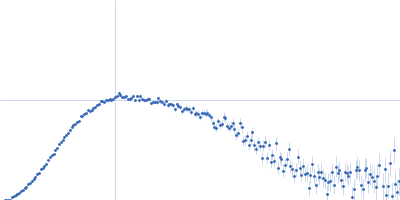
| Sample: |
Serine/arginine (SR)-type shuttling mRNA binding protein NPL3 monomer, 18 kDa Saccharomyces cerevisiae (strain … protein
Short RNA oligonucleotide (NPL3 binding sequence) monomer, 4 kDa RNA
|
| Buffer: |
20 mM NaPO4, 50 mM NaCl, 1 mM DTT, pH: 6.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, SFB 1035, Technische Universität München on 2020 Feb 20
|
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.3 |
nm |
| VolumePorod |
30 |
nm3 |
|
|