Asymmetric horseshoe-like assembly of peroxisomal yeast oxalyl-CoA synthetase.
Bürgi J,
Lill P,
Giannopoulou EA,
Jeffries CM,
Chojnowski G,
Raunser S,
Gatsogiannis C,
Wilmanns M
Biol Chem
(2023 Jan 26)
|
|
|
|
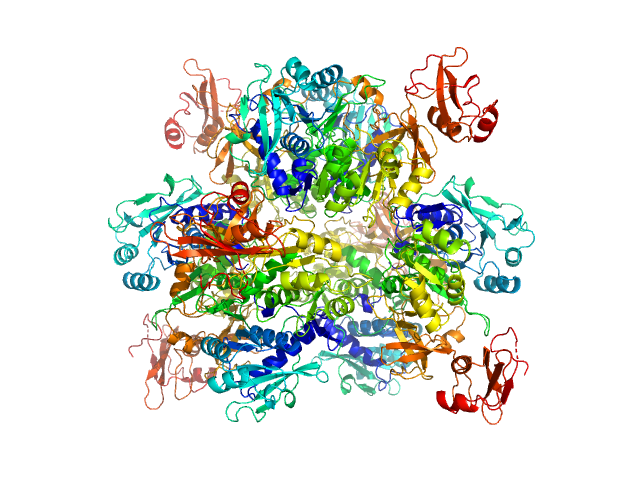
|
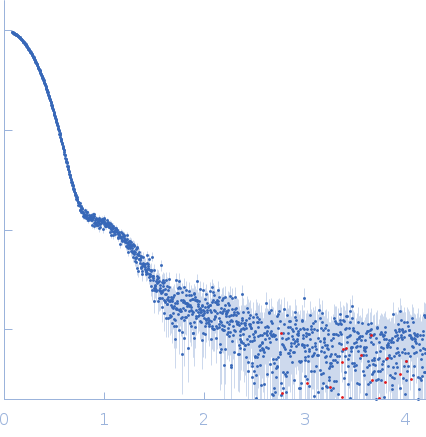
| Sample: |
Oxalate--CoA ligase, 363 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
50 mM Hepes, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Dec 18
|
|
| RgGuinier |
4.7 |
nm |
| Dmax |
13.2 |
nm |
| VolumePorod |
565 |
nm3 |
|
|
|
|
|
|
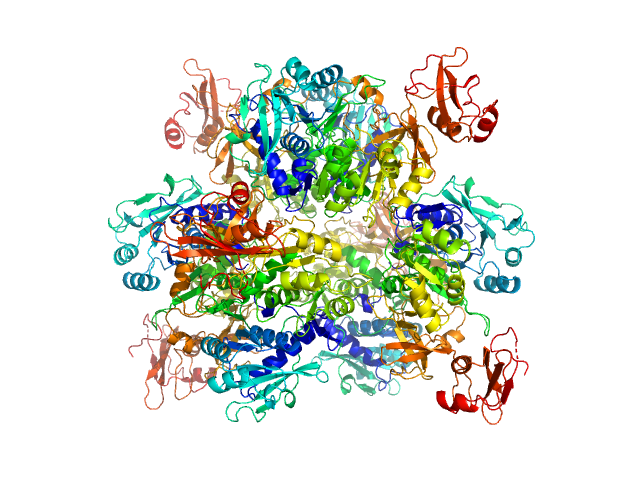
|
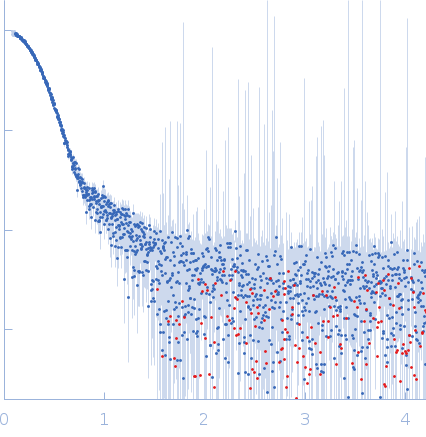
| Sample: |
Oxalate--CoA ligase, 363 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
50 mM Hepes, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Dec 18
|
|
| RgGuinier |
4.5 |
nm |
| Dmax |
13.7 |
nm |
| VolumePorod |
432 |
nm3 |
|
|
|
|
|
|
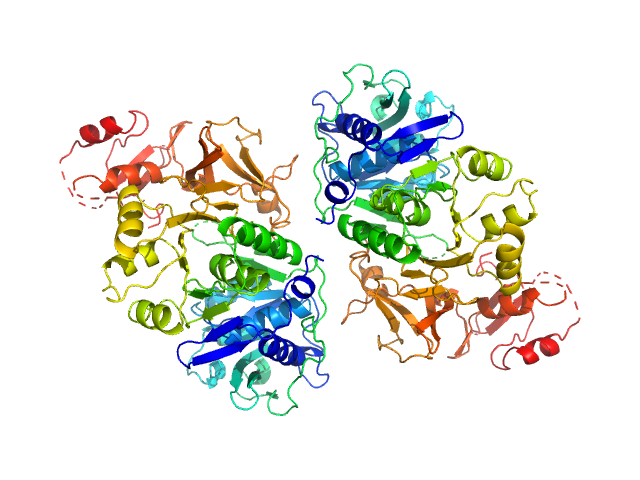
|
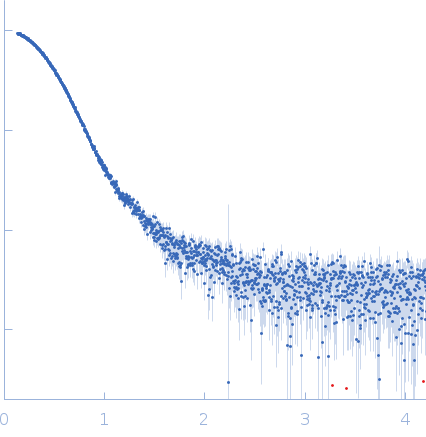
| Sample: |
Oxalate--CoA ligase (K352D) dimer, 121 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
50 mM Hepes, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Dec 18
|
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.1 |
nm |
| VolumePorod |
163 |
nm3 |
|
|
|
|
|
|
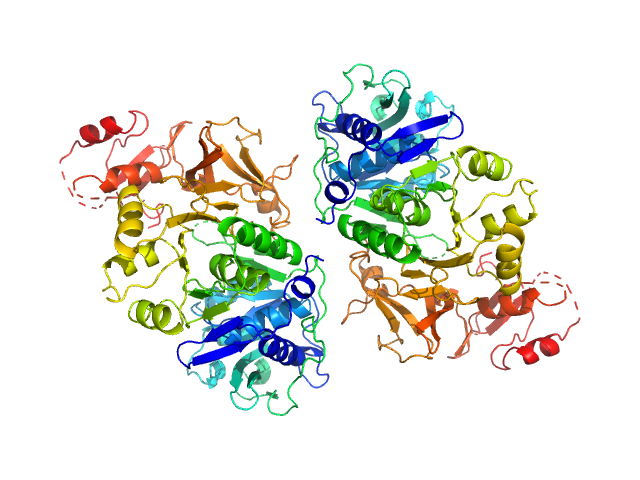
|
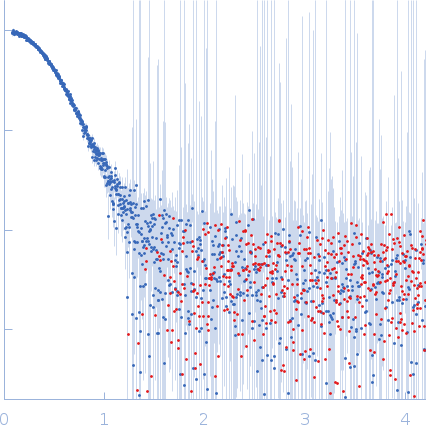
| Sample: |
Oxalate--CoA ligase (K352D) dimer, 121 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
50 mM Hepes, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Dec 18
|
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.2 |
nm |
| VolumePorod |
194 |
nm3 |
|
|