|
|
|
|
|
| Sample: |
Transient receptor potential cation channel subfamily V member 4 monomer, 15 kDa Gallus gallus protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, 10 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 16
|
Crosstalk between regulatory elements in disordered TRPV4 N-terminus modulates lipid-dependent channel activity
Nature Communications 14(1) (2023)
Goretzki B, Wiedemann C, McCray B, Schäfer S, Jansen J, Tebbe F, Mitrovic S, Nöth J, Cabezudo A, Donohue J, Jeffries C, Steinchen W, Stengel F, Sumner C, Hummer G, Hellmich U
|
| RgGuinier |
3.2 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Transient receptor potential cation channel subfamily V member 4 monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, 10 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 16
|
Crosstalk between regulatory elements in disordered TRPV4 N-terminus modulates lipid-dependent channel activity
Nature Communications 14(1) (2023)
Goretzki B, Wiedemann C, McCray B, Schäfer S, Jansen J, Tebbe F, Mitrovic S, Nöth J, Cabezudo A, Donohue J, Jeffries C, Steinchen W, Stengel F, Sumner C, Hummer G, Hellmich U
|
| RgGuinier |
3.3 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
37 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Rap guanine nucleotide exchange factor 3 monomer, 82 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 8.0, 150 mM NaCl, 0.5 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2017 Oct 4
|
Membranes prime the RapGEF EPAC1 to transduce cAMP signaling
Nature Communications 14(1) (2023)
Sartre C, Peurois F, Ley M, Kryszke M, Zhang W, Courilleau D, Fischmeister R, Ambroise Y, Zeghouf M, Cianferani S, Ferrandez Y, Cherfils J
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
130 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Rap guanine nucleotide exchange factor 3 monomer, 82 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 8.0, 150 mM NaCl, 0.5 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2017 Oct 4
|
Membranes prime the RapGEF EPAC1 to transduce cAMP signaling
Nature Communications 14(1) (2023)
Sartre C, Peurois F, Ley M, Kryszke M, Zhang W, Courilleau D, Fischmeister R, Ambroise Y, Zeghouf M, Cianferani S, Ferrandez Y, Cherfils J
|
| RgGuinier |
3.5 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
140 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Rap guanine nucleotide exchange factor 3 monomer, 82 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 8.0, 150 mM NaCl, 0.5 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2017 Oct 4
|
Membranes prime the RapGEF EPAC1 to transduce cAMP signaling
Nature Communications 14(1) (2023)
Sartre C, Peurois F, Ley M, Kryszke M, Zhang W, Courilleau D, Fischmeister R, Ambroise Y, Zeghouf M, Cianferani S, Ferrandez Y, Cherfils J
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.4 |
nm |
| VolumePorod |
140 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Rap guanine nucleotide exchange factor 3 monomer, 82 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 8.0, 150 mM NaCl, 0.5 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2017 Oct 4
|
Membranes prime the RapGEF EPAC1 to transduce cAMP signaling
Nature Communications 14(1) (2023)
Sartre C, Peurois F, Ley M, Kryszke M, Zhang W, Courilleau D, Fischmeister R, Ambroise Y, Zeghouf M, Cianferani S, Ferrandez Y, Cherfils J
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
134 |
nm3 |
|
|
|
|
|
|
|
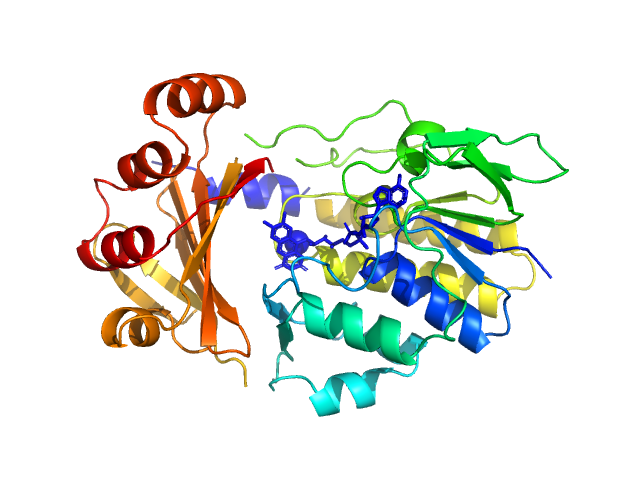
| Sample: |
Monooxygenase (M154I, A283T) monomer, 39 kDa Stenotrophomonas maltophilia protein
|
| Buffer: |
25 mM Bis-Tris, 150 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2022 Jan 10
|
Tetracycline-modifying enzyme Sm
TetX from Stenotrophomonas maltophilia
Acta Crystallographica Section F Structural Biology Communications 79(7):180-192 (2023)
Malý M, Kolenko P, Stránský J, Švecová L, Dušková J, Koval' T, Skálová T, Trundová M, Adámková K, Černý J, Božíková P, Dohnálek J
|
| RgGuinier |
2.4 |
nm |
| Dmax |
6.8 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
|
|
|
|
|
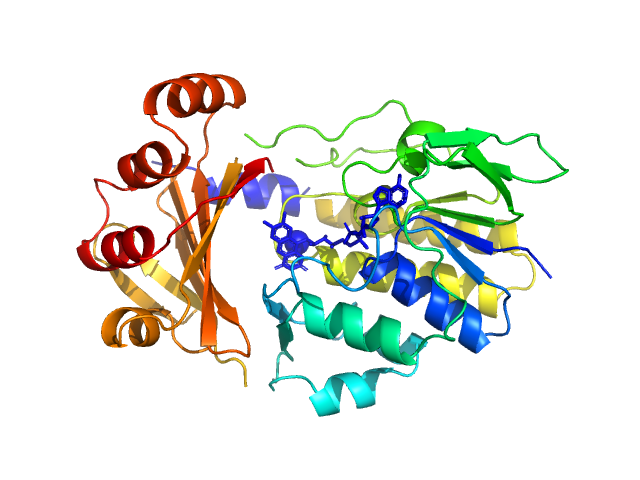
| Sample: |
Monooxygenase (M154I, A283T) monomer, 39 kDa Stenotrophomonas maltophilia protein
|
| Buffer: |
25 mM Bis-Tris, 150 mM NaCl, 30 mM DTT, pH: 6.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2022 Jan 7
|
Tetracycline-modifying enzyme Sm
TetX from Stenotrophomonas maltophilia
Acta Crystallographica Section F Structural Biology Communications 79(7):180-192 (2023)
Malý M, Kolenko P, Stránský J, Švecová L, Dušková J, Koval' T, Skálová T, Trundová M, Adámková K, Černý J, Božíková P, Dohnálek J
|
| RgGuinier |
2.4 |
nm |
| Dmax |
6.4 |
nm |
| VolumePorod |
64 |
nm3 |
|
|
|
|
|
|
|
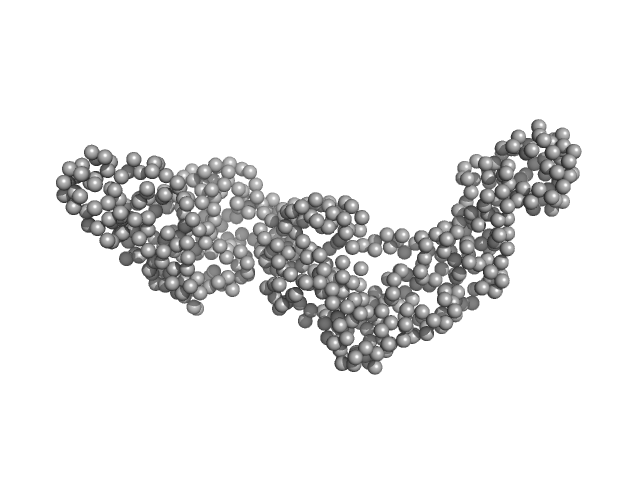
| Sample: |
Bartonella effector protein (Bep) substrate of VirB T4SS monomer, 64 kDa Bartonella clarridgeiae (strain … protein
|
| Buffer: |
25 mM Hepes, 300 mM NaCl, 1 mM TCEP, 5% v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at Xenocs Xeuss 2.0 Q-Xoom, Center for Structural Studies, Heinrich-Heine-University on 2020 May 29
|
Structure and function of Bartonella effector protein 1: target and interdomain interactions
University of Basel PhD thesis 15051 (2023)
Markus Huber, Jens Reiners
|
| RgGuinier |
4.1 |
nm |
| Dmax |
13.4 |
nm |
| VolumePorod |
83 |
nm3 |
|
|
|
|
|
|
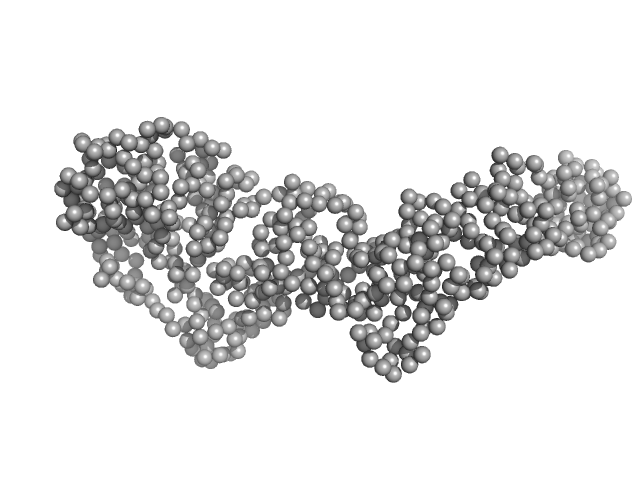
|
| Sample: |
Bartonella effector protein (Bep) substrate of VirB T4SS monomer, 64 kDa Bartonella clarridgeiae (strain … protein
|
| Buffer: |
25 mM Hepes, 300 mM NaCl, 1 mM TCEP, 5% v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 26
|
Structure and function of Bartonella effector protein 1: target and interdomain interactions
University of Basel PhD thesis 15051 (2023)
Markus Huber, Jens Reiners
|
| RgGuinier |
3.8 |
nm |
| Dmax |
13.4 |
nm |
| VolumePorod |
87 |
nm3 |
|
|