|
|
|
|
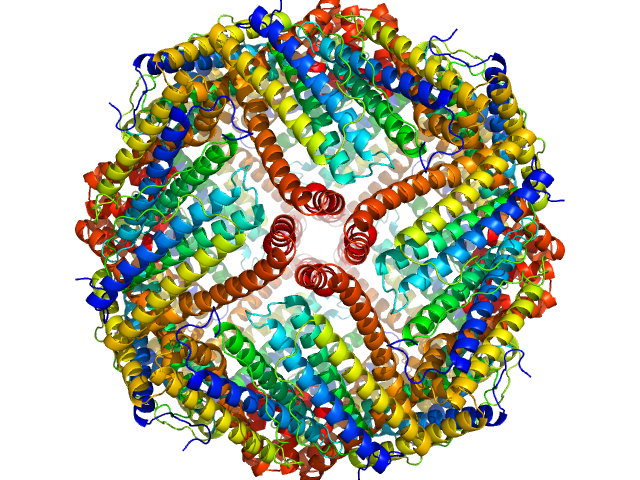
|
| Sample: |
Ferritin light chain 24-mer, 480 kDa Equus caballus protein
|
| Buffer: |
phosphate buffered saline, pH: 7.2
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Jun 19
|
AF4-to-SAXS: expanded characterization of nanoparticles and proteins at the P12 BioSAXS beamline.
J Synchrotron Radiat (2025)
Da Vela S, Bartels K, Franke D, Soloviov D, Gräwert T, Molodenskiy D, Kolb B, Wilhelmy C, Drexel R, Meier F, Haas H, Langguth P, Graewert MA
|
| RgGuinier |
5.4 |
nm |
| Dmax |
13.3 |
nm |
| VolumePorod |
632 |
nm3 |
|
|
|
|
|
|
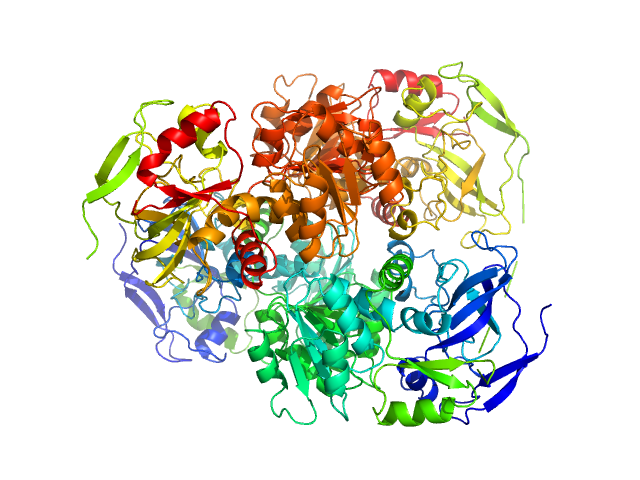
|
| Sample: |
Alcohol dehydrogenase 1 tetramer, 147 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
phosphate buffered saline, 1% glycerol, pH: 7.2
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Apr 13
|
AF4-to-SAXS: expanded characterization of nanoparticles and proteins at the P12 BioSAXS beamline.
J Synchrotron Radiat (2025)
Da Vela S, Bartels K, Franke D, Soloviov D, Gräwert T, Molodenskiy D, Kolb B, Wilhelmy C, Drexel R, Meier F, Haas H, Langguth P, Graewert MA
|
| RgGuinier |
3.4 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
197 |
nm3 |
|
|
|
|
|
|
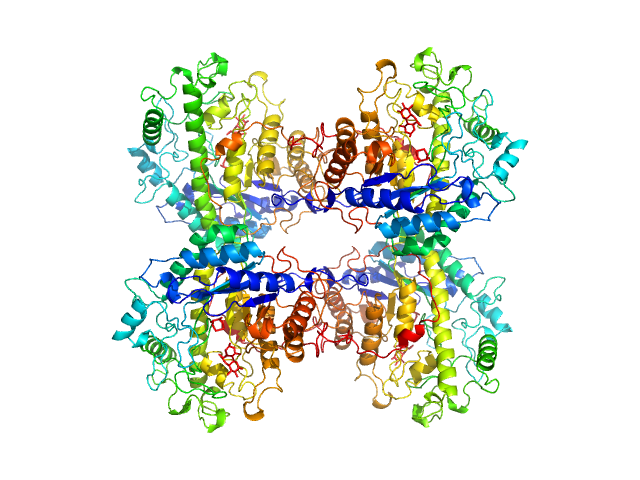
|
| Sample: |
Beta-amylase tetramer, 224 kDa Ipomoea batatas protein
|
| Buffer: |
phosphate buffered saline, 1% glycerol, pH: 7.2
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Apr 13
|
AF4-to-SAXS: expanded characterization of nanoparticles and proteins at the P12 BioSAXS beamline.
J Synchrotron Radiat (2025)
Da Vela S, Bartels K, Franke D, Soloviov D, Gräwert T, Molodenskiy D, Kolb B, Wilhelmy C, Drexel R, Meier F, Haas H, Langguth P, Graewert MA
|
| RgGuinier |
4.2 |
nm |
| Dmax |
12.6 |
nm |
| VolumePorod |
297 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Sigma-Aldrich polystyrene nanospheres with a diameter of 100 nm monomer, 0 kDa synthetic construct
|
| Buffer: |
water/detergent solution (0.0125 % (v/v) NovaChem), pH: 7
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Jun 19
|
AF4-to-SAXS: expanded characterization of nanoparticles and proteins at the P12 BioSAXS beamline.
J Synchrotron Radiat (2025)
Da Vela S, Bartels K, Franke D, Soloviov D, Gräwert T, Molodenskiy D, Kolb B, Wilhelmy C, Drexel R, Meier F, Haas H, Langguth P, Graewert MA
|
| RgGuinier |
47.7 |
nm |
| Dmax |
111.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
Thermo Fisher polystyrene nanospheres with a diameter of 20 nm monomer, 1 kDa synthetic construct
|
| Buffer: |
water/detergent solution (0.0125 % (v/v) NovaChem), pH: 7
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Jun 19
|
AF4-to-SAXS: expanded characterization of nanoparticles and proteins at the P12 BioSAXS beamline.
J Synchrotron Radiat (2025)
Da Vela S, Bartels K, Franke D, Soloviov D, Gräwert T, Molodenskiy D, Kolb B, Wilhelmy C, Drexel R, Meier F, Haas H, Langguth P, Graewert MA
|
| RgGuinier |
9.0 |
nm |
| Dmax |
25.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Precursor microRNA 20a monomer, 23 kDa Homo sapiens RNA
|
| Buffer: |
50 mM potassium phosphate buffer, 1 mM MgCl2, 50 mM NaCl, 5 mM BME, pH: 7.5
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Oct 8
|
Structural features within precursor microRNA-20a regulate Dicer-TRBP processing
Journal of Molecular Biology :169317 (2025)
Liu Y, Harkner C, Westwood M, Munsayac A, Keane S
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Splicing Factor Proline/Glutamine rich (1-598) dimer, 130 kDa Homo sapiens protein
|
| Buffer: |
500mM KNO3, 20mM HEPES, 5% glycerol, 1mM DTT, pH: 7.4
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2023 Mar 13
|
Structural dynamics of IDR interactions in human SFPQ and implications for liquid–liquid phase separation
Acta Crystallographica Section D Structural Biology 81(7):357-379 (2025)
Koning H, Lai V, Sethi A, Chakraborty S, Ang C, Fox A, Duff A, Whitten A, Marshall A, Bond C
|
| RgGuinier |
7.6 |
nm |
| Dmax |
34.3 |
nm |
| VolumePorod |
400 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Splicing factor, proline- and glutamine-rich dimer, 153 kDa Homo sapiens protein
|
| Buffer: |
150 mM KCl, 0.075% glycerol, 20 mM HEPES, 0.5 mM DTT,, pH: 7.4
|
| Experiment: |
SANS
data collected at Quokka - Small Angle Neutron Scattering, Australian Centre for Neutron Scattering (ANSTO) on 2023 Jan 20
|
Structural dynamics of IDR interactions in human SFPQ and implications for liquid–liquid phase separation
Acta Crystallographica Section D Structural Biology 81(7):357-379 (2025)
Koning H, Lai V, Sethi A, Chakraborty S, Ang C, Fox A, Duff A, Whitten A, Marshall A, Bond C
|
| RgGuinier |
6.1 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
181 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Splicing factor, proline- and glutamine-rich dimer, 153 kDa Homo sapiens protein
|
| Buffer: |
150 mM KCl, 1.5% glycerol, 20 mM HEPES, 1 mM DTT (in 100% H2O), pH: 7.4
|
| Experiment: |
SANS
data collected at Quokka - Small Angle Neutron Scattering, Australian Centre for Neutron Scattering (ANSTO) on 2023 Jan 23
|
Structural dynamics of IDR interactions in human SFPQ and implications for liquid–liquid phase separation
Acta Crystallographica Section D Structural Biology 81(7):357-379 (2025)
Koning H, Lai V, Sethi A, Chakraborty S, Ang C, Fox A, Duff A, Whitten A, Marshall A, Bond C
|
| RgGuinier |
8.7 |
nm |
| Dmax |
30.7 |
nm |
| VolumePorod |
241000 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Splicing factor, proline- and glutamine-rich dimer, 153 kDa Homo sapiens protein
|
| Buffer: |
500mM KNO3, 20mM HEPES, 5% glycerol, 1mM DTT, pH: 7.4
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2022 Oct 22
|
Structural dynamics of IDR interactions in human SFPQ and implications for liquid–liquid phase separation
Acta Crystallographica Section D Structural Biology 81(7):357-379 (2025)
Koning H, Lai V, Sethi A, Chakraborty S, Ang C, Fox A, Duff A, Whitten A, Marshall A, Bond C
|
| RgGuinier |
7.9 |
nm |
| Dmax |
42.5 |
nm |
| VolumePorod |
485 |
nm3 |
|
|