|
|
|
|
|
| Sample: |
Ferredoxin protease E83A mutant monomer, 101 kDa Pectobacterium atrosepticum SCRI1043 protein
Arabidopsis ferredoxin 2 monomer, 11 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Oct 26
|
FusC, a member of the M16 protease family acquired by bacteria for iron piracy against plants.
PLoS Biol 16(8):e2006026 (2018)
Grinter R, Hay ID, Song J, Wang J, Teng D, Dhanesakaran V, Wilksch JJ, Davies MR, Littler D, Beckham SA, Henderson IR, Strugnell RA, Dougan G, Lithgow T
|
| RgGuinier |
3.7 |
nm |
| Dmax |
13.6 |
nm |
| VolumePorod |
149 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ferredoxin protease E83A mutant monomer, 101 kDa Pectobacterium atrosepticum SCRI1043 protein
Arabidopsis ferredoxin 2 monomer, 11 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Oct 26
|
FusC, a member of the M16 protease family acquired by bacteria for iron piracy against plants.
PLoS Biol 16(8):e2006026 (2018)
Grinter R, Hay ID, Song J, Wang J, Teng D, Dhanesakaran V, Wilksch JJ, Davies MR, Littler D, Beckham SA, Henderson IR, Strugnell RA, Dougan G, Lithgow T
|
| RgGuinier |
3.7 |
nm |
| Dmax |
13.6 |
nm |
| VolumePorod |
155 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ferredoxin protease E83A mutant monomer, 101 kDa Pectobacterium atrosepticum SCRI1043 protein
Arabidopsis ferredoxin 2 monomer, 11 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Oct 26
|
FusC, a member of the M16 protease family acquired by bacteria for iron piracy against plants.
PLoS Biol 16(8):e2006026 (2018)
Grinter R, Hay ID, Song J, Wang J, Teng D, Dhanesakaran V, Wilksch JJ, Davies MR, Littler D, Beckham SA, Henderson IR, Strugnell RA, Dougan G, Lithgow T
|
| RgGuinier |
3.7 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
156 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ferredoxin protease E83A mutant monomer, 101 kDa Pectobacterium atrosepticum SCRI1043 protein
Arabidopsis ferredoxin 2 monomer, 11 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Oct 26
|
FusC, a member of the M16 protease family acquired by bacteria for iron piracy against plants.
PLoS Biol 16(8):e2006026 (2018)
Grinter R, Hay ID, Song J, Wang J, Teng D, Dhanesakaran V, Wilksch JJ, Davies MR, Littler D, Beckham SA, Henderson IR, Strugnell RA, Dougan G, Lithgow T
|
| RgGuinier |
3.8 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
160 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ferredoxin protease E83A mutant monomer, 101 kDa Pectobacterium atrosepticum SCRI1043 protein
Arabidopsis ferredoxin 2 monomer, 11 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Oct 26
|
FusC, a member of the M16 protease family acquired by bacteria for iron piracy against plants.
PLoS Biol 16(8):e2006026 (2018)
Grinter R, Hay ID, Song J, Wang J, Teng D, Dhanesakaran V, Wilksch JJ, Davies MR, Littler D, Beckham SA, Henderson IR, Strugnell RA, Dougan G, Lithgow T
|
| RgGuinier |
3.7 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
165 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ferredoxin protease E83A mutant monomer, 101 kDa Pectobacterium atrosepticum SCRI1043 protein
Arabidopsis ferredoxin 2 monomer, 11 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Oct 26
|
FusC, a member of the M16 protease family acquired by bacteria for iron piracy against plants.
PLoS Biol 16(8):e2006026 (2018)
Grinter R, Hay ID, Song J, Wang J, Teng D, Dhanesakaran V, Wilksch JJ, Davies MR, Littler D, Beckham SA, Henderson IR, Strugnell RA, Dougan G, Lithgow T
|
| RgGuinier |
3.7 |
nm |
| Dmax |
13.7 |
nm |
| VolumePorod |
177 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ferredoxin protease E83A mutant monomer, 101 kDa Pectobacterium atrosepticum SCRI1043 protein
Arabidopsis ferredoxin 2 monomer, 11 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Oct 26
|
FusC, a member of the M16 protease family acquired by bacteria for iron piracy against plants.
PLoS Biol 16(8):e2006026 (2018)
Grinter R, Hay ID, Song J, Wang J, Teng D, Dhanesakaran V, Wilksch JJ, Davies MR, Littler D, Beckham SA, Henderson IR, Strugnell RA, Dougan G, Lithgow T
|
| RgGuinier |
3.8 |
nm |
| Dmax |
14.2 |
nm |
| VolumePorod |
178 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ferredoxin protease E83A mutant monomer, 101 kDa Pectobacterium atrosepticum SCRI1043 protein
Arabidopsis ferredoxin 2 monomer, 11 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Oct 26
|
FusC, a member of the M16 protease family acquired by bacteria for iron piracy against plants.
PLoS Biol 16(8):e2006026 (2018)
Grinter R, Hay ID, Song J, Wang J, Teng D, Dhanesakaran V, Wilksch JJ, Davies MR, Littler D, Beckham SA, Henderson IR, Strugnell RA, Dougan G, Lithgow T
|
| RgGuinier |
3.7 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
175 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ferredoxin protease E83A mutant monomer, 101 kDa Pectobacterium atrosepticum SCRI1043 protein
Arabidopsis ferredoxin 2 monomer, 11 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Oct 26
|
FusC, a member of the M16 protease family acquired by bacteria for iron piracy against plants.
PLoS Biol 16(8):e2006026 (2018)
Grinter R, Hay ID, Song J, Wang J, Teng D, Dhanesakaran V, Wilksch JJ, Davies MR, Littler D, Beckham SA, Henderson IR, Strugnell RA, Dougan G, Lithgow T
|
| RgGuinier |
3.7 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
180 |
nm3 |
|
|
|
|
|
|
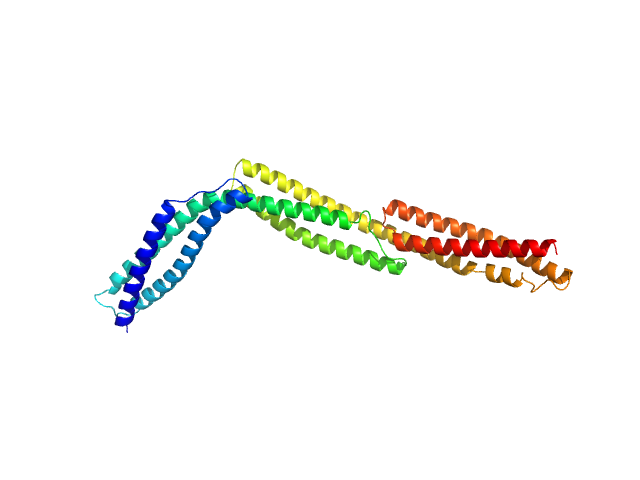
|
| Sample: |
R1-3 human dystrophin fragment monomer, 39 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-d11, 150 mM NaCl, 0.1 mM EDTA-d16, in 100% D2O, pD 7.5, pH: 7.1 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2016 Nov 7
|
Human Dystrophin Structural Changes upon Binding to Anionic Membrane Lipids.
Biophys J 115(7):1231-1239 (2018)
Dos Santos Morais R, Delalande O, Pérez J, Mias-Lucquin D, Lagarrigue M, Martel A, Molza AE, Chéron A, Raguénès-Nicol C, Chenuel T, Bondon A, Appavou MS, Le Rumeur E, Combet S, Hubert JF
|
| RgGuinier |
4.2 |
nm |
| Dmax |
17.7 |
nm |
| VolumePorod |
46 |
nm3 |
|
|