|
|
|
|

|
| Sample: |
Peroxisomal targeting signal 1 receptor monomer, 71 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES-KOH (pH 7.5), 100mM KCl, and 20mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Feb 24
|
Solution structure of human Pex5.Pex14.PTS1 protein complexes obtained by small angle X-ray scattering.
J Biol Chem 284(37):25334-42 (2009)
Shiozawa K, Konarev PV, Neufeld C, Wilmanns M, Svergun DI
|
| RgGuinier |
5.0 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
181 |
nm3 |
|
|
|
|
|
|
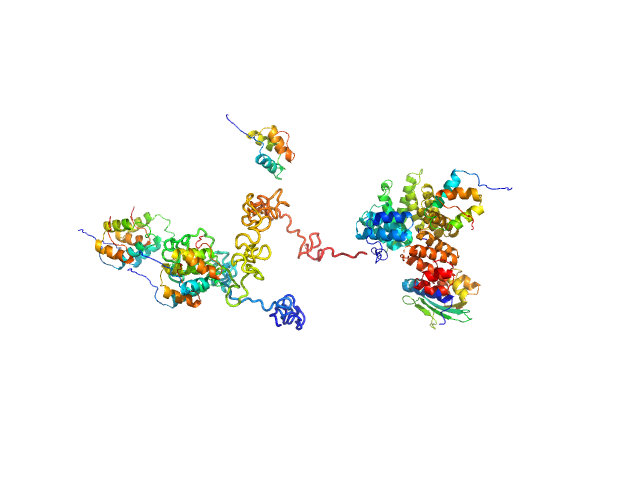
|
| Sample: |
Peroxisomal targeting signal 1 receptor monomer, 71 kDa Homo sapiens protein
Peroxisomal membrane protein PEX14 monomer, 7 kDa Homo sapiens protein
PTS1-BP monomer, 13 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES-KOH (pH 7.5), 100mM KCl, and 20mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Mar 18
|
Solution structure of human Pex5.Pex14.PTS1 protein complexes obtained by small angle X-ray scattering.
J Biol Chem 284(37):25334-42 (2009)
Shiozawa K, Konarev PV, Neufeld C, Wilmanns M, Svergun DI
|
| RgGuinier |
6.0 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
267 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Isoform 3 of Rap guanine nucleotide exchange factor 4 monomer, 114 kDa Mus musculus protein
|
| Buffer: |
150 mM NaCl, 1 mM EDTA, 1 mM DTT, and 10 mM Tris-HCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSess, University of Utah on 2008 Aug 7
|
Mechanism of Epac activation: structural and functional analyses of Epac2 hinge mutants with constitutive and reduced activities.
J Biol Chem 284(35):23644-51 (2009)
Tsalkova T, Blumenthal DK, Mei FC, White MA, Cheng X
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
123 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Isoform 3 of Rap guanine nucleotide exchange factor 4 monomer, 114 kDa Mus musculus protein
|
| Buffer: |
150 mM NaCl, 1 mM EDTA, 1 mM DTT, and 10 mM Tris-HCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSess, University of Utah on 2008 Aug 8
|
Mechanism of Epac activation: structural and functional analyses of Epac2 hinge mutants with constitutive and reduced activities.
J Biol Chem 284(35):23644-51 (2009)
Tsalkova T, Blumenthal DK, Mei FC, White MA, Cheng X
|
| RgGuinier |
3.8 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
151 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Isoform 3 of Rap guanine nucleotide exchange factor 4 monomer, 114 kDa Mus musculus protein
|
| Buffer: |
150 mM NaCl, 1 mM EDTA, 1 mM DTT, and 10 mM Tris-HCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSess, University of Utah on 2008 Aug 11
|
Mechanism of Epac activation: structural and functional analyses of Epac2 hinge mutants with constitutive and reduced activities.
J Biol Chem 284(35):23644-51 (2009)
Tsalkova T, Blumenthal DK, Mei FC, White MA, Cheng X
|
| RgGuinier |
3.6 |
nm |
| Dmax |
11.4 |
nm |
| VolumePorod |
145 |
nm3 |
|
|
|
|
|
|
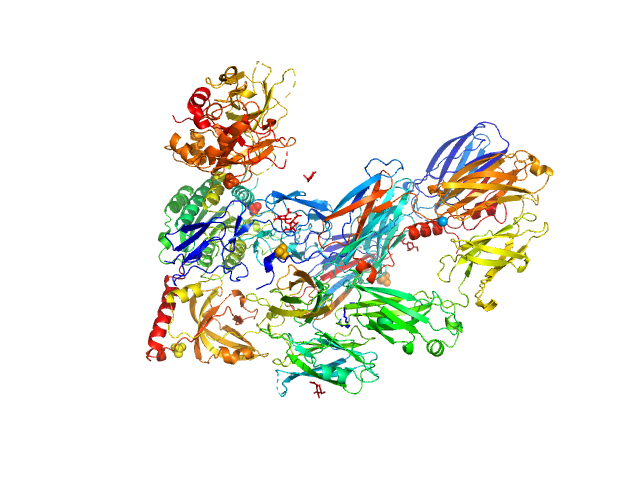
|
| Sample: |
Cobra venom factor monomer, 185 kDa Naja kaouthia protein
|
| Buffer: |
10 mM Tris 5 mM MgCl2 10 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Dec 19
|
Insights into complement convertase formation based on the structure of the factor B-cobra venom factor complex
The EMBO Journal 28(16):2469-2478 (2009)
Janssen B, Gomes L, Koning R, Svergun D, Koster A, Fritzinger D, Vogel C, Gros P
|
| RgGuinier |
4.6 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
384 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
ProNGF dimer, 45 kDa Mouse submandibulary glands protein
|
| Buffer: |
50 mM Naphosphat 0.5 M Ammonium Sulfate(NH4)2SO4, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Oct 25
|
Intrinsic structural disorder of mouse proNGF.
Proteins 75(4):990-1009 (2009)
Paoletti F, Covaceuszach S, Konarev PV, Gonfloni S, Malerba F, Schwarz E, Svergun DI, Cattaneo A, Lamba D
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
92 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Endoglin tetramer, 243 kDa Homo sapiens protein
|
| Buffer: |
10 mM Tris–HCl, 50 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2008 Oct 17
|
Structural and functional characterization of soluble endoglin receptor
Biochemical and Biophysical Research Communications 383(4):386-391 (2009)
Le B, Franke D, Svergun D, Han T, Hwang H, Kim K
|
| RgGuinier |
7.6 |
nm |
| Dmax |
26.0 |
nm |
| VolumePorod |
434 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Endoglin dimer, 121 kDa Homo sapiens protein
|
| Buffer: |
10 mM Tris–HCl, 50 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2008 Oct 17
|
Structural and functional characterization of soluble endoglin receptor
Biochemical and Biophysical Research Communications 383(4):386-391 (2009)
Le B, Franke D, Svergun D, Han T, Hwang H, Kim K
|
| RgGuinier |
4.7 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
173 |
nm3 |
|
|
|
|
|
|
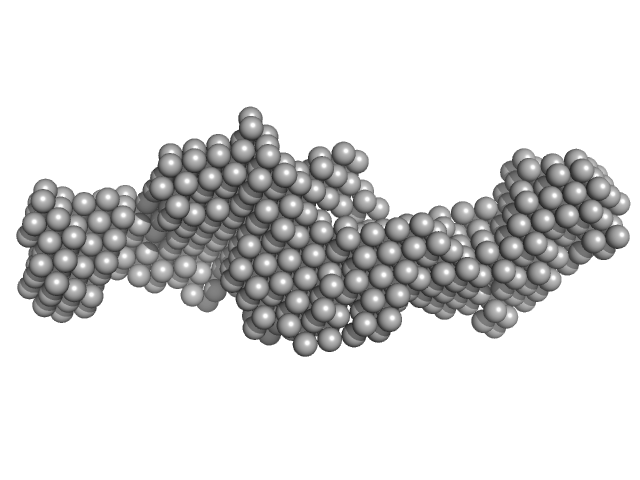
|
| Sample: |
Nuclear fragile X mental retardation-interacting protein 2 monomer, 59 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl pH 8.0, 5 mM TCEP, pH: |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Oct 11
|
A study of the ultrastructure of fragile-X-related proteins.
Biochem J 419(2):347-57 (2009)
Sjekloća L, Konarev PV, Eccleston J, Taylor IA, Svergun DI, Pastore A
|
| RgGuinier |
4.1 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
92 |
nm3 |
|
|