|
|
|
|
|
| Sample: |
Sorting nexin-9 monomer, 19 kDa Homo sapiens protein
|
| Buffer: |
137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, 3 %Glycerol, pH: 8.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Mar 20
|
The Chlamydia pneumoniae effector SemD exploits its host’s endocytic machinery by structural and functional mimicry
Nature Communications 15(1) (2024)
Kocher F, Applegate V, Reiners J, Port A, Spona D, Hänsch S, Mirzaiebadizi A, Ahmadian M, Smits S, Hegemann J, Mölleken K
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.1 |
nm |
| VolumePorod |
42 |
nm3 |
|
|
|
|
|
|
|
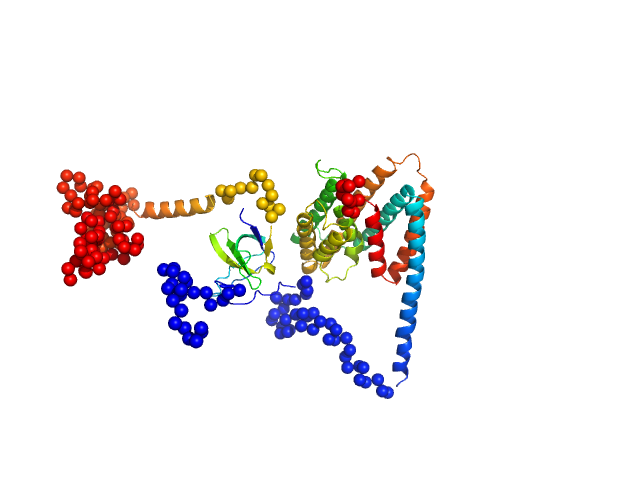
| Sample: |
Uncharacterized protein monomer, 35 kDa Chlamydia pneumoniae protein
Sorting nexin-9 monomer, 19 kDa Homo sapiens protein
|
| Buffer: |
137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, 3 %Glycerol, pH: 8.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Mar 20
|
The Chlamydia pneumoniae effector SemD exploits its host’s endocytic machinery by structural and functional mimicry
Nature Communications 15(1) (2024)
Kocher F, Applegate V, Reiners J, Port A, Spona D, Hänsch S, Mirzaiebadizi A, Ahmadian M, Smits S, Hegemann J, Mölleken K
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
94 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Uncharacterized protein monomer, 35 kDa Chlamydia pneumoniae protein
Actin nucleation-promoting factor WASL monomer, 19 kDa Rattus norvegicus protein
|
| Buffer: |
137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, 3 %Glycerol, pH: 8.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Dec 14
|
The Chlamydia pneumoniae effector SemD exploits its host’s endocytic machinery by structural and functional mimicry
Nature Communications 15(1) (2024)
Kocher F, Applegate V, Reiners J, Port A, Spona D, Hänsch S, Mirzaiebadizi A, Ahmadian M, Smits S, Hegemann J, Mölleken K
|
| RgGuinier |
4.2 |
nm |
| Dmax |
15.9 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Protein W monomer, 6 kDa Hendra virus (isolate … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2023 Apr 16
|
Dissecting Henipavirus W proteins conformational and fibrillation properties: contribution of their N- and C-terminal constituent domains.
FEBS J (2024)
Pesce G, Gondelaud F, Ptchelkine D, Bignon C, Fourquet P, Longhi S
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
8 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Protein W monomer, 6 kDa Henipavirus nipahense protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2023 Apr 16
|
Dissecting Henipavirus W proteins conformational and fibrillation properties: contribution of their N- and C-terminal constituent domains.
FEBS J (2024)
Pesce G, Gondelaud F, Ptchelkine D, Bignon C, Fourquet P, Longhi S
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
8 |
nm3 |
|
|
|
|
|
|
|
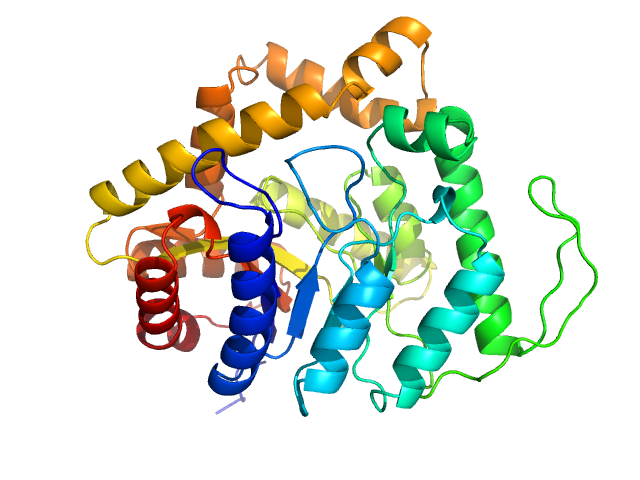
| Sample: |
Alkanal monooxygenase alpha chain dimer, 84 kDa Enhygromyxa salina protein
|
| Buffer: |
150 mM NaCl, 25 mM Tris, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2020 Aug 1
|
luxA
Gene From Enhygromyxa salina
Encodes a Functional Homodimeric Luciferase
Proteins: Structure, Function, and Bioinformatics (2024)
Yudenko A, Bazhenov S, Aleksenko V, Goncharov I, Semenov O, Remeeva A, Nazarenko V, Kuznetsova E, Fomin V, Konopleva M, Al Ebrahim R, Sluchanko N, Ryzhykau Y, Semenov Y, Kuklin A, Manukhov I, Gushchin I
|
| RgGuinier |
3.3 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
138 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
E3 ubiquitin-protein ligase LRSAM1 monomer, 84 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris-HCl pH 8.0, 150 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2019 Dec 7
|
Leucine Rich Repeat And Sterile Alpha Motif Containing 1 (LRSAM1)
Si Hoon Park
|
| RgGuinier |
6.1 |
nm |
| Dmax |
19.4 |
nm |
| VolumePorod |
250 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
E3 ubiquitin-protein ligase LRSAM1 - ΔPTAP/PSAP-RING monomer, 74 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl pH 8.0, 300 mM NaCl, 1 mM TCEP, 5% w/v glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 4C, Pohang Accelerator Laboratory on 2017 Oct 24
|
Leucine Rich Repeat And Sterile Alpha Motif Containing 1 (LRSAM1)
Si Hoon Park
|
| RgGuinier |
5.5 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
322 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
E3 ubiquitin-protein ligase LRSAM1 - LRR containing N-terminal domain monomer, 35 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl pH 8.0, 300 mM NaCl, 1 mM TCEP, 5% w/v glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 4C, Pohang Accelerator Laboratory on 2017 Oct 23
|
Leucine Rich Repeat And Sterile Alpha Motif Containing 1 (LRSAM1)
Si Hoon Park
|
| RgGuinier |
3.4 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
105 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
E3 ubiquitin-protein ligase LRSAM1 - SAM domain monomer, 8 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at 4C, Pohang Accelerator Laboratory on 2019 Apr 4
|
Leucine Rich Repeat And Sterile Alpha Motif Containing 1 (LRSAM1)
Si Hoon Park
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.3 |
nm |
| VolumePorod |
11 |
nm3 |
|
|