|
|
|
|
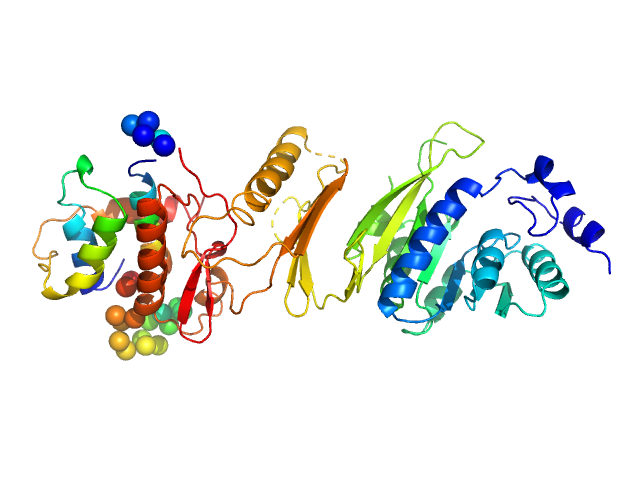
|
| Sample: |
Bifunctional kinase- methyltransferase WbdD monomer, 59 kDa Escherichia coli protein
|
| Buffer: |
20 mM BisTris 50 mM NaCl 5 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Sep 23
|
A coiled-coil domain acts as a molecular ruler to regulate O-antigen chain length in lipopolysaccharide.
Nat Struct Mol Biol 22(1):50-56 (2015)
Hagelueken G, Clarke BR, Huang H, Tuukkanen A, Danciu I, Svergun DI, Hussain R, Liu H, Whitfield C, Naismith JH
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
90 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Alpha domain of Ag43a monomer, 49 kDa Escherichia coli protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2009 Nov 19
|
The antigen 43 structure reveals a molecular Velcro-like mechanism of autotransporter-mediated bacterial clumping.
Proc Natl Acad Sci U S A 111(1):457-62 (2014)
Heras B, Totsika M, Peters KM, Paxman JJ, Gee CL, Jarrott RJ, Perugini MA, Whitten AE, Schembri MA
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
62 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
DNA mismatch repair protein MutS dimer, 191 kDa Escherichia coli protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Feb 28
|
Using stable MutS dimers and tetramers to quantitatively analyze DNA mismatch recognition and sliding clamp formation.
Nucleic Acids Res 41(17):8166-81 (2013)
Groothuizen FS, Fish A, Petoukhov MV, Reumer A, Manelyte L, Winterwerp HH, Marinus MG, Lebbink JH, Svergun DI, Friedhoff P, Sixma TK
|
| RgGuinier |
4.7 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
307 |
nm3 |
|
|
|
|
|
|
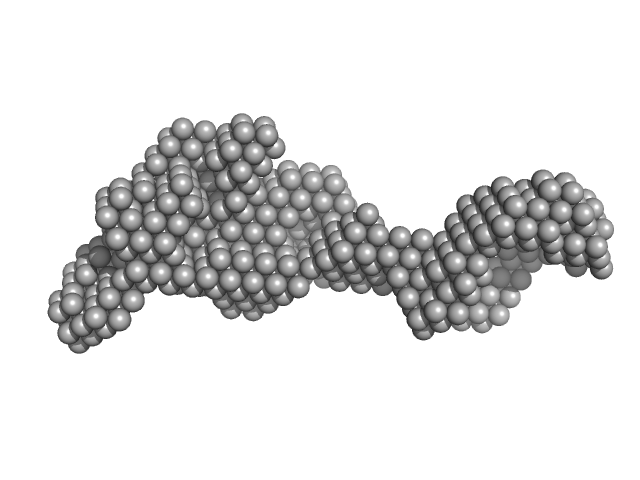
|
| Sample: |
DNA mismatch repair protein MutS tetramer, 381 kDa Escherichia coli protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 May 12
|
Using stable MutS dimers and tetramers to quantitatively analyze DNA mismatch recognition and sliding clamp formation.
Nucleic Acids Res 41(17):8166-81 (2013)
Groothuizen FS, Fish A, Petoukhov MV, Reumer A, Manelyte L, Winterwerp HH, Marinus MG, Lebbink JH, Svergun DI, Friedhoff P, Sixma TK
|
| RgGuinier |
7.8 |
nm |
| Dmax |
28.0 |
nm |
| VolumePorod |
700 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA mismatch repair protein MutS tetramer, 381 kDa Escherichia coli protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 May 12
|
Using stable MutS dimers and tetramers to quantitatively analyze DNA mismatch recognition and sliding clamp formation.
Nucleic Acids Res 41(17):8166-81 (2013)
Groothuizen FS, Fish A, Petoukhov MV, Reumer A, Manelyte L, Winterwerp HH, Marinus MG, Lebbink JH, Svergun DI, Friedhoff P, Sixma TK
|
| RgGuinier |
7.8 |
nm |
| Dmax |
27.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
DNA mismatch repair protein MutS tetramer, 381 kDa Escherichia coli protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 May 12
|
Using stable MutS dimers and tetramers to quantitatively analyze DNA mismatch recognition and sliding clamp formation.
Nucleic Acids Res 41(17):8166-81 (2013)
Groothuizen FS, Fish A, Petoukhov MV, Reumer A, Manelyte L, Winterwerp HH, Marinus MG, Lebbink JH, Svergun DI, Friedhoff P, Sixma TK
|
| RgGuinier |
8.5 |
nm |
| Dmax |
29.0 |
nm |
| VolumePorod |
750 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA mismatch repair protein MutS tetramer, 381 kDa Escherichia coli protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 May 12
|
Using stable MutS dimers and tetramers to quantitatively analyze DNA mismatch recognition and sliding clamp formation.
Nucleic Acids Res 41(17):8166-81 (2013)
Groothuizen FS, Fish A, Petoukhov MV, Reumer A, Manelyte L, Winterwerp HH, Marinus MG, Lebbink JH, Svergun DI, Friedhoff P, Sixma TK
|
| RgGuinier |
8.3 |
nm |
| Dmax |
29.0 |
nm |
| VolumePorod |
720 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA mismatch repair protein MutS tetramer, 381 kDa Escherichia coli protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 May 12
|
Using stable MutS dimers and tetramers to quantitatively analyze DNA mismatch recognition and sliding clamp formation.
Nucleic Acids Res 41(17):8166-81 (2013)
Groothuizen FS, Fish A, Petoukhov MV, Reumer A, Manelyte L, Winterwerp HH, Marinus MG, Lebbink JH, Svergun DI, Friedhoff P, Sixma TK
|
|
|
|
|
|
|
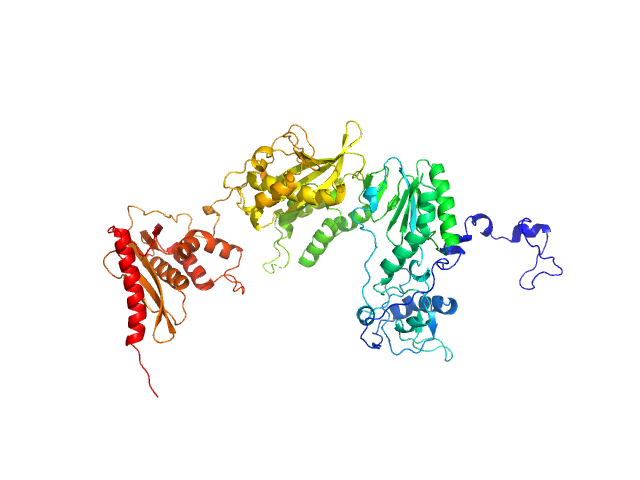
|
| Sample: |
Apo XMRV RT monomer, 75 kDa Escherichia coli protein
|
| Buffer: |
10 mM HEPES 100 mM KCl 5% Glycerol, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Dec 8
|
Structural analysis of monomeric retroviral reverse transcriptase in complex with an RNA/DNA hybrid.
Nucleic Acids Res 41(6):3874-87 (2013)
Nowak E, Potrzebowski W, Konarev PV, Rausch JW, Bona MK, Svergun DI, Bujnicki JM, Le Grice SF, Nowotny M
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
160 |
nm3 |
|
|
|
|
|
|
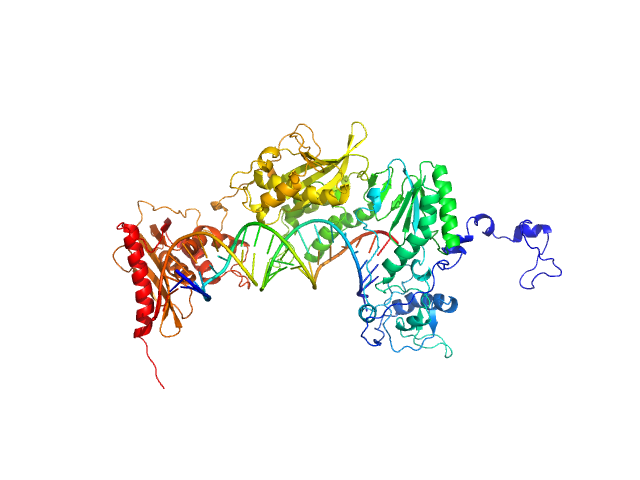
|
| Sample: |
Apo XMRV RT monomer, 75 kDa Escherichia coli protein
RNA_DNA hybrid substrate monomer, 15 kDa
|
| Buffer: |
10 mM HEPES 100 mM KCl 5% Glycerol, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Dec 8
|
Structural analysis of monomeric retroviral reverse transcriptase in complex with an RNA/DNA hybrid.
Nucleic Acids Res 41(6):3874-87 (2013)
Nowak E, Potrzebowski W, Konarev PV, Rausch JW, Bona MK, Svergun DI, Bujnicki JM, Le Grice SF, Nowotny M
|
| RgGuinier |
3.5 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
155 |
nm3 |
|
|