|
|
|
|
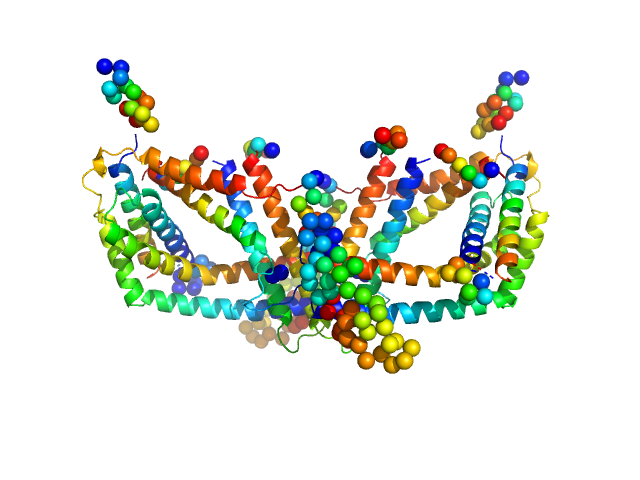
|
| Sample: |
Polyphosphate-targeting protein A dimer, 79 kDa Streptomyces chartreusis protein
|
| Buffer: |
20 mM Tris-HCl 400 mM NaCl, pH: 7.4
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Nov 24
|
Structural and biochemical analysis of a phosin from Streptomyces chartreusis reveals a combined polyphosphate- and metal-binding fold.
FEBS Lett (2019)
Werten S, Rustmeier NH, Gemmer M, Virolle MJ, Hinrichs W
|
| RgGuinier |
3.5 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
124 |
nm3 |
|
|
|
|
|
|
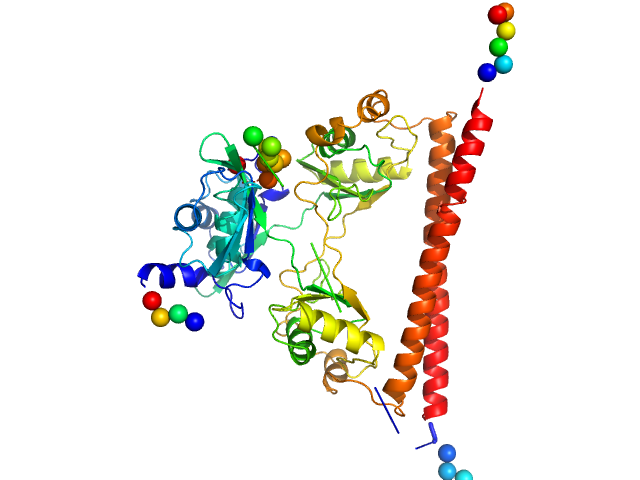
|
| Sample: |
Splicing factor, proline- and glutamine-rich dimer, 60 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 250 mM NaCl, 5% (v/v) glycerol, pH: 7.5
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Apr 19
|
A new crystal structure and small-angle X-ray scattering analysis of the homodimer of human SFPQ.
Acta Crystallogr F Struct Biol Commun 75(Pt 6):439-449 (2019)
Hewage TW, Caria S, Lee M
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
91 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA protection during starvation protein dodecamer, 224 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM Tris-HCl, 50 mM NaCl, 0.5 mM EDTA, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Nov 27
|
Protective Dps-DNA co-crystallization in stressed cells: an in vitro structural study by small-angle X-ray scattering and cryo-electron tomography.
FEBS Lett 593(12):1360-1371 (2019)
Dadinova LA, Chesnokov YM, Kamyshinsky RA, Orlov IA, Petoukhov MV, Mozhaev AA, Soshinskaya EY, Lazarev VN, Manuvera VA, Orekhov AS, Vasiliev AL, Shtykova EV
|
|
|
|
|
|
|
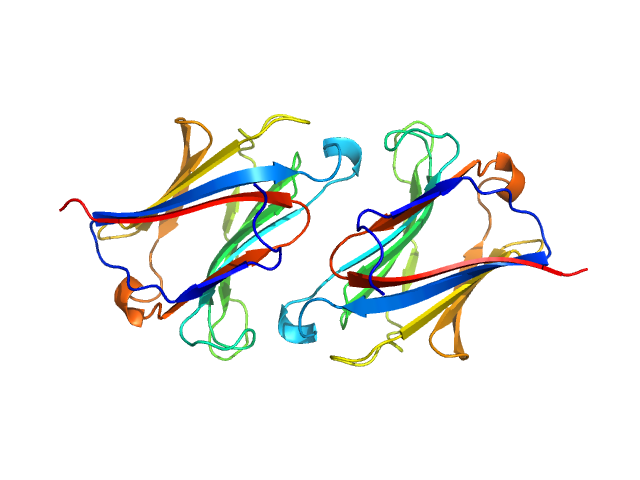
|
| Sample: |
Galectin-10 Tyr69Glu dimer, 33 kDa Homo sapiens protein
|
| Buffer: |
20 mM Hepes 150 NaCl, pH: 7.4
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2018 Feb 4
|
Protein crystallization promotes type 2 immunity and is reversible by antibody treatment.
Science 364(6442) (2019)
Persson EK, Verstraete K, Heyndrickx I, Gevaert E, Aegerter H, Percier JM, Deswarte K, Verschueren KHG, Dansercoer A, Gras D, Chanez P, Bachert C, Gonçalves A, Van Gorp H, De Haard H, Blanchetot C, Sa...
|
| RgGuinier |
2.1 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nuclear receptor CoRepressor 1; Nuclear Receptor Interaction Domain (NID) monomer, 29 kDa Mus musculus protein
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, 2 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Jun 20
|
Interplay of Protein Disorder in Retinoic Acid Receptor Heterodimer and Its Corepressor Regulates Gene Expression.
Structure (2019)
Cordeiro TN, Sibille N, Germain P, Barthe P, Boulahtouf A, Allemand F, Bailly R, Vivat V, Ebel C, Barducci A, Bourguet W, le Maire A, Bernadó P
|
| RgGuinier |
4.7 |
nm |
| Dmax |
17.7 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nuclear receptor CoRepressor 1; Nuclear Receptor Interaction Domain (NID) monomer, 29 kDa Mus musculus protein
Retinoid-X receptor alpha (RXR-alpha) Ligand Binding Domain (LBD) monomer, 26 kDa Mus musculus protein
Retinoic acid receptor alpha (RAR-alpha) Ligand binding domain (LDB) monomer, 28 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, 2 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Jul 23
|
Interplay of Protein Disorder in Retinoic Acid Receptor Heterodimer and Its Corepressor Regulates Gene Expression.
Structure (2019)
Cordeiro TN, Sibille N, Germain P, Barthe P, Boulahtouf A, Allemand F, Bailly R, Vivat V, Ebel C, Barducci A, Bourguet W, le Maire A, Bernadó P
|
| RgGuinier |
4.8 |
nm |
| Dmax |
19.4 |
nm |
| VolumePorod |
167 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nuclear receptor CoRepressor 1; Nuclear Receptor Interaction Domain (NID) monomer, 29 kDa Mus musculus protein
Retinoid-X receptor alpha (RXR-alpha) Ligand Binding Domain (LBD) monomer, 26 kDa Mus musculus protein
Retinoid-X receptor alpha (RXR-alpha) Δ helix12 monomer, 24 kDa Mus musculus protein
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, 2 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Jul 23
|
Interplay of Protein Disorder in Retinoic Acid Receptor Heterodimer and Its Corepressor Regulates Gene Expression.
Structure (2019)
Cordeiro TN, Sibille N, Germain P, Barthe P, Boulahtouf A, Allemand F, Bailly R, Vivat V, Ebel C, Barducci A, Bourguet W, le Maire A, Bernadó P
|
| RgGuinier |
4.2 |
nm |
| Dmax |
15.7 |
nm |
| VolumePorod |
183 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nuclear receptor CoRepressor 1; Nuclear Receptor Interaction Domain (NID) monomer, 29 kDa Mus musculus protein
Retinoid-X receptor alpha (RXR-alpha) Ligand Binding Domain (LBD) monomer, 26 kDa Mus musculus protein
Retinoic acid receptor alpha (RAR-alpha) Ligand binding domain (LDB) monomer, 28 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, 2 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Jul 23
|
Interplay of Protein Disorder in Retinoic Acid Receptor Heterodimer and Its Corepressor Regulates Gene Expression.
Structure (2019)
Cordeiro TN, Sibille N, Germain P, Barthe P, Boulahtouf A, Allemand F, Bailly R, Vivat V, Ebel C, Barducci A, Bourguet W, le Maire A, Bernadó P
|
| RgGuinier |
4.8 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
178 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nuclear receptor CoRepressor 1; Nuclear Receptor Interaction Domain (NID) monomer, 29 kDa Mus musculus protein
Retinoid-X receptor alpha (RXR-alpha) Ligand Binding Domain (LBD) monomer, 26 kDa Mus musculus protein
Retinoic acid receptor alpha (RAR-alpha) Ligand binding domain (LDB) mutant I396E monomer, 28 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, 2 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Mar 9
|
Interplay of Protein Disorder in Retinoic Acid Receptor Heterodimer and Its Corepressor Regulates Gene Expression.
Structure (2019)
Cordeiro TN, Sibille N, Germain P, Barthe P, Boulahtouf A, Allemand F, Bailly R, Vivat V, Ebel C, Barducci A, Bourguet W, le Maire A, Bernadó P
|
| RgGuinier |
5.3 |
nm |
| Dmax |
22.4 |
nm |
| VolumePorod |
171 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nuclear receptor CoRepressor 1; Nuclear Receptor Interaction Domain (NID) monomer, 29 kDa Mus musculus protein
Retinoid-X receptor alpha (RXR-alpha) Ligand Binding Domain (LBD) monomer, 26 kDa Mus musculus protein
Retinoic acid receptor alpha (RAR-alpha) Ligand binding domain (LDB) monomer, 28 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, 2 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Jul 23
|
Interplay of Protein Disorder in Retinoic Acid Receptor Heterodimer and Its Corepressor Regulates Gene Expression.
Structure (2019)
Cordeiro TN, Sibille N, Germain P, Barthe P, Boulahtouf A, Allemand F, Bailly R, Vivat V, Ebel C, Barducci A, Bourguet W, le Maire A, Bernadó P
|
| RgGuinier |
4.2 |
nm |
| Dmax |
17.2 |
nm |
| VolumePorod |
131 |
nm3 |
|
|