|
|
|
|

|
| Sample: |
Integrin beta-4 monomer, 23 kDa Homo sapiens protein
|
| Buffer: |
20 mM Sodium Phosphate 150 mM NaCl 5% glycerol 3 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Aug 6
|
Combination of X-ray crystallography, SAXS and DEER to obtain the structure of the FnIII-3,4 domains of integrin α6β4.
Acta Crystallogr D Biol Crystallogr 71(Pt 4):969-85 (2015)
Alonso-García N, García-Rubio I, Manso JA, Buey RM, Urien H, Sonnenberg A, Jeschke G, de Pereda JM
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
|
|
|
|
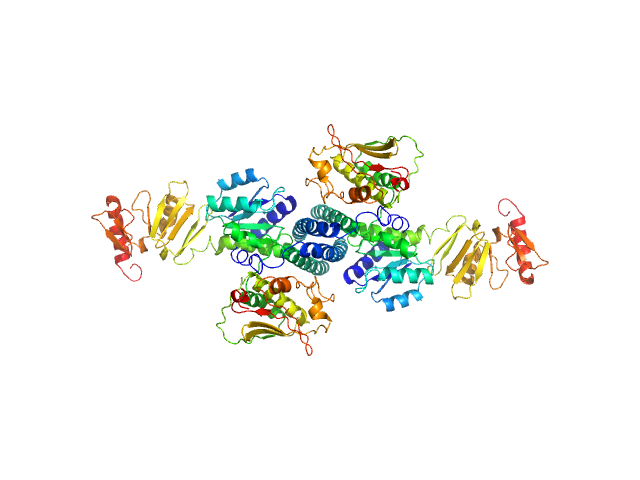
|
| Sample: |
Histidine protein kinase dimer, 54 kDa Streptococcus pneumoniae protein
Response regulator dimer, 61 kDa Streptococcus pneumoniae protein
|
| Buffer: |
20 mM Tris 200 mM NaCl 5% (v/v) Glycerol 5 mM β-mercaptoethanol, pH: 7.5
|
| Experiment: |
SAXS
data collected at Bruker Nanostar, IBBMC on 2012 May 16
|
Modeling the ComD/ComE/comcde interaction network using small angle X-ray scattering.
FEBS J 282(8):1538-53 (2015)
Sanchez D, Boudes M, van Tilbeurgh H, Durand D, Quevillon-Cheruel S
|
| RgGuinier |
4.0 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
175 |
nm3 |
|
|
|
|
|
|
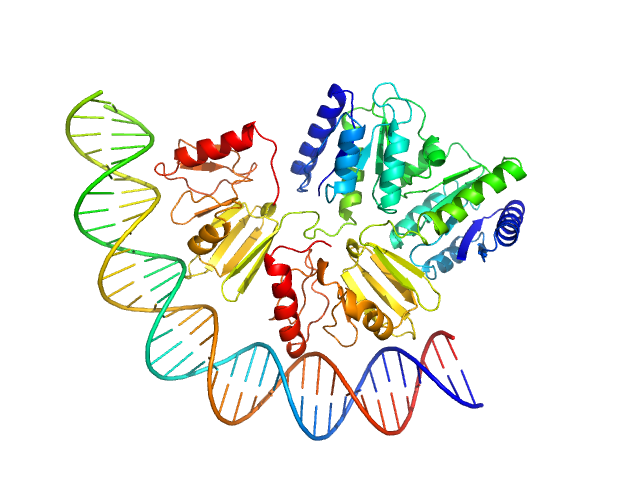
|
| Sample: |
comcde , 24 kDa Streptococcus pneumoniae DNA
Response regulator dimer, 61 kDa Streptococcus pneumoniae protein
|
| Buffer: |
50 mM MES 500 mM NaCl 5% (v/v) Glycerol 5 mM β-mercaptoethanol, pH: 6.2
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2011 Feb 11
|
Modeling the ComD/ComE/comcde interaction network using small angle X-ray scattering.
FEBS J 282(8):1538-53 (2015)
Sanchez D, Boudes M, van Tilbeurgh H, Durand D, Quevillon-Cheruel S
|
| RgGuinier |
3.4 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
122 |
nm3 |
|
|
|
|
|
|
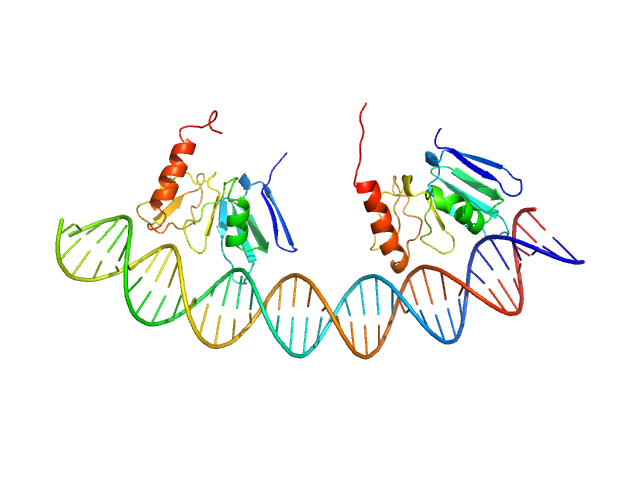
|
| Sample: |
comcde , 24 kDa Streptococcus pneumoniae DNA
Response regulator dimer, 29 kDa Streptococcus pneumoniae protein
|
| Buffer: |
50 mM MES 500 mM NaCl 5% (v/v) Glycerol 5 mM β-mercaptoethanol, pH: 6.2
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2013 Jun 2
|
Modeling the ComD/ComE/comcde interaction network using small angle X-ray scattering.
FEBS J 282(8):1538-53 (2015)
Sanchez D, Boudes M, van Tilbeurgh H, Durand D, Quevillon-Cheruel S
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
74 |
nm3 |
|
|
|
|
|
|
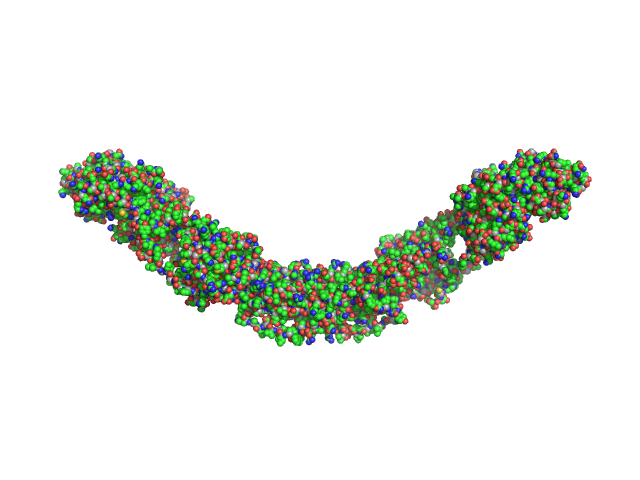
|
| Sample: |
Maltose Binding Protein fused to Protein Interacting with C kinase 1 dimer, 174 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris 300 mM NaCl 1 mM maltose 1 mM EGTA 2 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 13
|
PICK1 is implicated in organelle motility in an Arp2/3 complex-independent manner.
Mol Biol Cell 26(7):1308-22 (2015)
Madasu Y, Yang C, Boczkowska M, Bethoney KA, Zwolak A, Rebowski G, Svitkina T, Dominguez R
|
| RgGuinier |
8.4 |
nm |
| Dmax |
27.6 |
nm |
| VolumePorod |
483 |
nm3 |
|
|
|
|
|
|
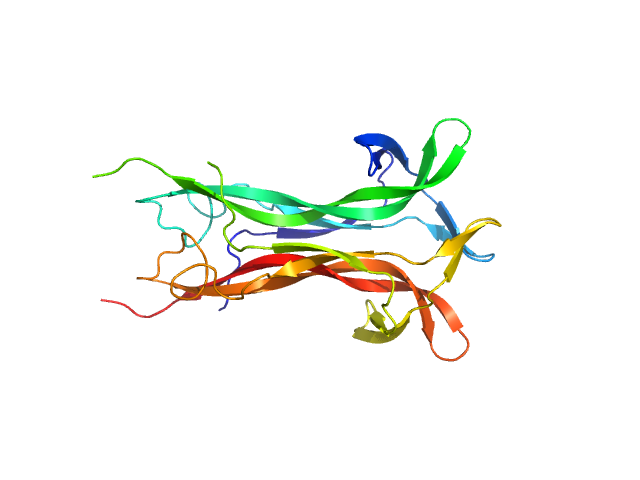
|
| Sample: |
Beta-nerve growth factor dimer, 24 kDa Homo sapiens protein
|
| Buffer: |
50 mM Na-phosphate, 1 mM EDTA, pH: 7
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Jul 2
|
The conundrum of the high-affinity NGF binding site formation unveiled?
Biophys J 108(3):687-97 (2015)
Covaceuszach S, Konarev PV, Cassetta A, Paoletti F, Svergun DI, Lamba D, Cattaneo A
|
|
|
|
|
|
|
|
| Sample: |
Ethylene Receptor 1 dimer, 129 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM Tris-NDSB 150 mM NaCl 1mM DTT 250 mM NDSB, pH: 8.8
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Mar 19
|
Structural model of the cytosolic domain of the plant ethylene receptor 1 (ETR1).
J Biol Chem 290(5):2644-58 (2015)
Mayerhofer H, Panneerselvam S, Kaljunen H, Tuukkanen A, Mertens HD, Mueller-Dieckmann J
|
| RgGuinier |
4.7 |
nm |
| Dmax |
15.8 |
nm |
| VolumePorod |
316 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ethylene Receptor 1 dimer, 129 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM TRIS 150 mM NaCl 1mM DTT 250mM NDSB, pH: 8.8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Mar 29
|
Structural model of the cytosolic domain of the plant ethylene receptor 1 (ETR1).
J Biol Chem 290(5):2644-58 (2015)
Mayerhofer H, Panneerselvam S, Kaljunen H, Tuukkanen A, Mertens HD, Mueller-Dieckmann J
|
| RgGuinier |
5.5 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
274 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ethylene receptor 1 dimerization histidine phosphotransfer + catalytic ATP-binding + receiver domains dimer, 88 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 1 mM DTT 5 mM ADP, pH: 8.8
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Oct 11
|
Structural model of the cytosolic domain of the plant ethylene receptor 1 (ETR1).
J Biol Chem 290(5):2644-58 (2015)
Mayerhofer H, Panneerselvam S, Kaljunen H, Tuukkanen A, Mertens HD, Mueller-Dieckmann J
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.9 |
nm |
| VolumePorod |
144 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ethylene receptor 1 dimerization histidine phosphotransfer + catalytic ATP-binding domains dimer, 55 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 1 mM DTT 5 mM ADP, pH: 8.8
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Oct 11
|
Structural model of the cytosolic domain of the plant ethylene receptor 1 (ETR1).
J Biol Chem 290(5):2644-58 (2015)
Mayerhofer H, Panneerselvam S, Kaljunen H, Tuukkanen A, Mertens HD, Mueller-Dieckmann J
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.7 |
nm |
| VolumePorod |
74 |
nm3 |
|
|