|
|
|
|
|
| Sample: |
humanized immunoglobulin G4 monoclonal antibody (lebrikizumab) monomer, 148 kDa Mouse/human protein
|
| Buffer: |
100 mM glycine-HCl, 200 mM NaCl, pH: 2
|
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2024 Nov 19
|
IgG4
and IgG1
undergo common acid‐induced compaction into an alternatively folded state
FEBS Letters (2025)
Imamura H, Honda S
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
238 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
humanized immunoglobulin G1 monoclonal antibody (trastuzumab) monomer, 148 kDa Mouse/human protein
|
| Buffer: |
100 mM glycine-HCl, 200 mM NaCl, pH: 2
|
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2023 May 20
|
IgG4
and IgG1
undergo common acid‐induced compaction into an alternatively folded state
FEBS Letters (2025)
Imamura H, Honda S
|
| RgGuinier |
4.2 |
nm |
| Dmax |
12.9 |
nm |
| VolumePorod |
240 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
humanized immunoglobulin G4 monoclonal antibody (lebrikizumab) monomer, 148 kDa Mouse/human protein
|
| Buffer: |
10 mM sodium phosphate, pH: 7
|
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2024 Nov 19
|
IgG4
and IgG1
undergo common acid‐induced compaction into an alternatively folded state
FEBS Letters (2025)
Imamura H, Honda S
|
| RgGuinier |
4.8 |
nm |
| Dmax |
15.6 |
nm |
| VolumePorod |
211 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
LNA-DNA-LNA monomer, 4 kDa DNA
|
| Buffer: |
phosphate buffered saline, pH: 7.4
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Oct 21
|
Application of enhanced biophysical strategies to determine ASO/target RNA binding affinity and kinetics
Michael Lerche
|
| RgGuinier |
1.3 |
nm |
| Dmax |
4.0 |
nm |
| VolumePorod |
4 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
MOE PS gapmers 3-8-3 monomer, 4 kDa DNA
|
| Buffer: |
phosphate buffered saline, pH: 7.4
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Oct 21
|
Application of enhanced biophysical strategies to determine ASO/target RNA binding affinity and kinetics
Michael Lerche
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.3 |
nm |
| VolumePorod |
5 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
oME ASO monomer, 4 kDa RNA
|
| Buffer: |
phosphate buffered saline, pH: 7.4
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Oct 21
|
Application of enhanced biophysical strategies to determine ASO/target RNA binding affinity and kinetics
Michael Lerche
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.0 |
nm |
| VolumePorod |
7 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
LNA-DNA-LNA monomer, 4 kDa DNA
PSCK9, 24mer ASO binding site monomer, 8 kDa RNA
|
| Buffer: |
phosphate buffered saline, pH: 7.4
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Oct 21
|
Application of enhanced biophysical strategies to determine ASO/target RNA binding affinity and kinetics
Michael Lerche
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
15 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
MOE PS gapmers 3-8-3 monomer, 4 kDa DNA
PSCK9, 24mer ASO binding site monomer, 8 kDa RNA
|
| Buffer: |
phosphate buffered saline, pH: 7.4
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Oct 21
|
Application of enhanced biophysical strategies to determine ASO/target RNA binding affinity and kinetics
Michael Lerche
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.4 |
nm |
| VolumePorod |
16 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
oME ASO monomer, 4 kDa RNA
PSCK9, 24mer ASO binding site monomer, 8 kDa RNA
|
| Buffer: |
phosphate buffered saline, pH: 7.4
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Oct 21
|
Application of enhanced biophysical strategies to determine ASO/target RNA binding affinity and kinetics
Michael Lerche
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
16 |
nm3 |
|
|
|
|
|
|
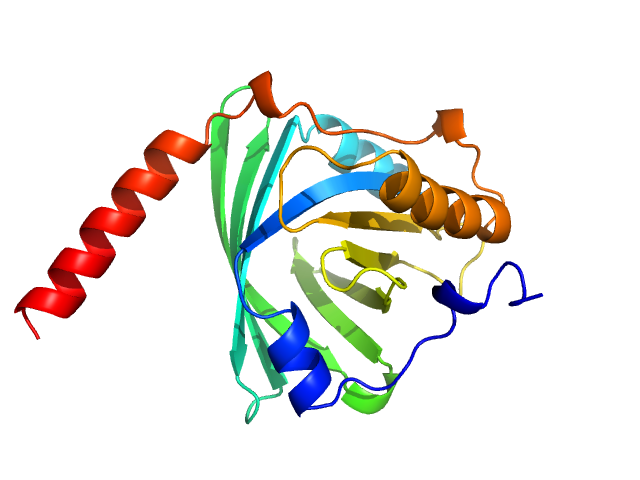
|
| Sample: |
Alpha-1-acid glycoprotein 1 monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
phosphate buffered saline, pH: 7.4
|
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2021 Jan 12
|
SAXS data based glycosylated models of human alpha-1-acid glycorprotein, a key player in health, disease and drug circulation.
J Biomol Struct Dyn :1-15 (2025)
Kalidas N, Peddada N, Pandey K, Ashish
|
| RgGuinier |
2.6 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
83 |
nm3 |
|
|