|
Synchrotron SAXS
data from solutions of
Protein Interacting with C-kinase 1 (PICK1) LKV, dimer contribution (data decomposition).
in
50 mM Tris 125 mM NaCl 0.01 vol% reduced TX-100, pH 7.4
were collected
on the
EMBL X33 beam line
at the DORIS III, DESY storage ring
(Hamburg, Germany)
using a MAR 345 Image Plate detector
(I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle).
Solute concentrations ranging between 4.2 and 8.8 mg/ml were measured
at 4°C.
Four successive
40 second frames were collected.
The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
The low angle data collected at lower concentration were merged with the highest concentration high angle data to yield the final composite scattering curve.
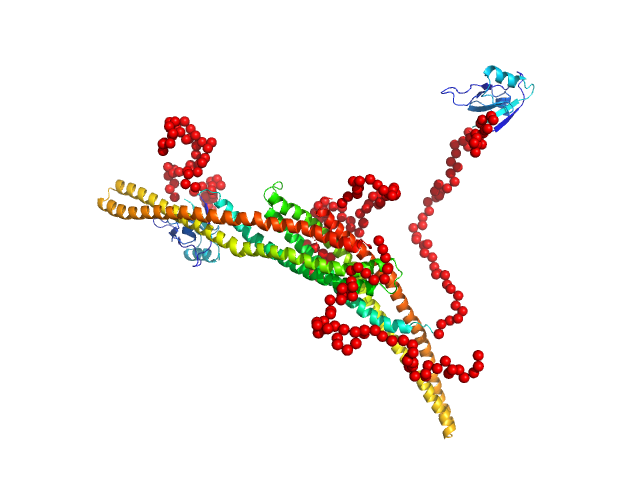
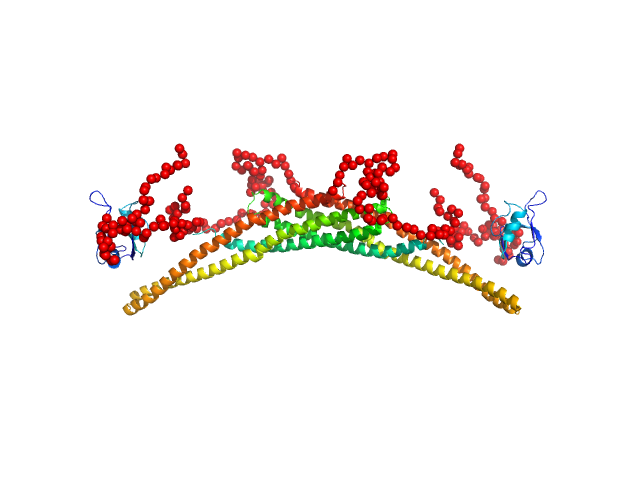
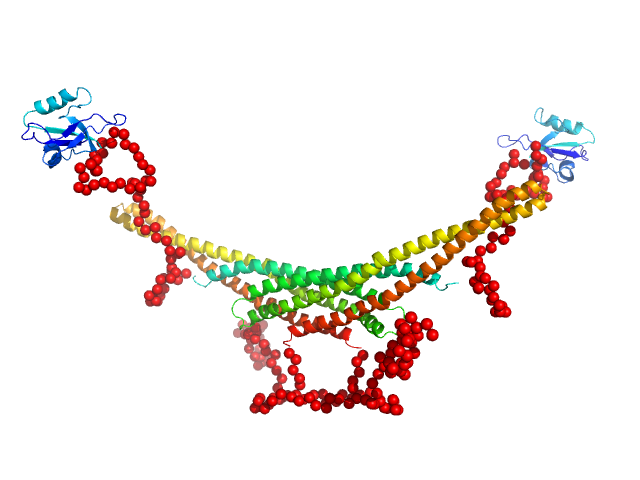
Structure and flexibility of PICK1 LKV mutant dimer was characterized by SAXS using data obtained by decomposing scattering from two polydisperse samples into dimer (and tetramer) contributions. To obtain the dimer scattering, two samples were measured at 4.4 and 8,8 mg/ml, and decomposed as described in the reference. Models were fitted to data using a combination of rigid body modelling and EOM, based on structural components defined by NMR and MD simulations. PDB files represent structures found in the optimal ensemble.
All data, in addition to that displayed for this entry, are available in the .zip archive.
|
|
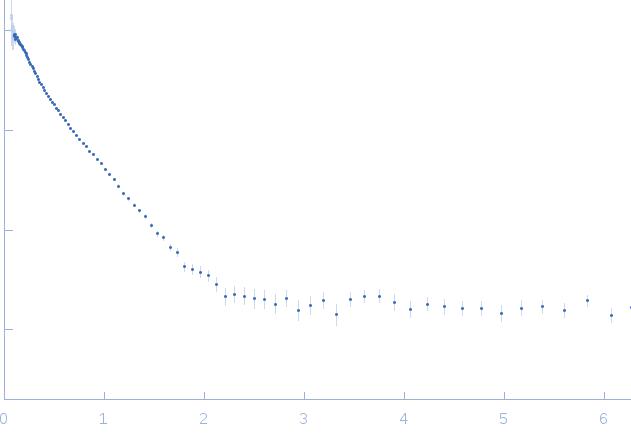 s, nm-1
s, nm-1
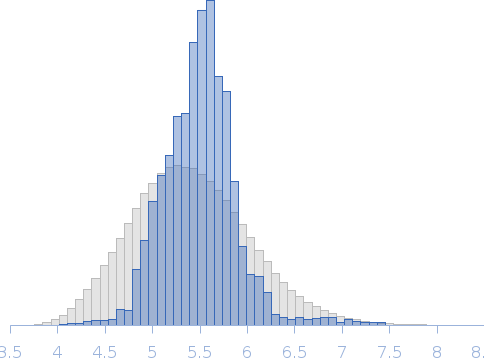 Rg, nm
Rg, nm