| MWexperimental | 143 | kDa |
| MWexpected | 246 | kDa |
| VPorod | 240 | nm3 |
|
log I(s)
2.50×10-2
2.50×10-3
2.50×10-4
2.50×10-5
|
 s, nm-1
s, nm-1
|
|
|
|
|
|
|
|
|
|
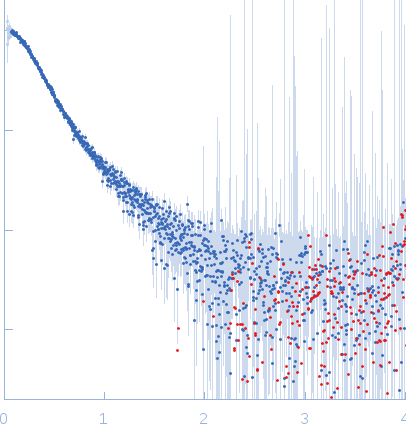
Synchrotron SAXS
data from solutions of
IL-6R tetramer-dimer equilibrium
in
water, pH 7.5
were collected
on the
B21 beam line
at the Diamond Light Source storage ring
(Didcot, UK)
using a Pilatus 2M detector
at a sample-detector distance of 3.9 m and
(I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle).
One solute concentration of 0.10 mg/ml was measured.
The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
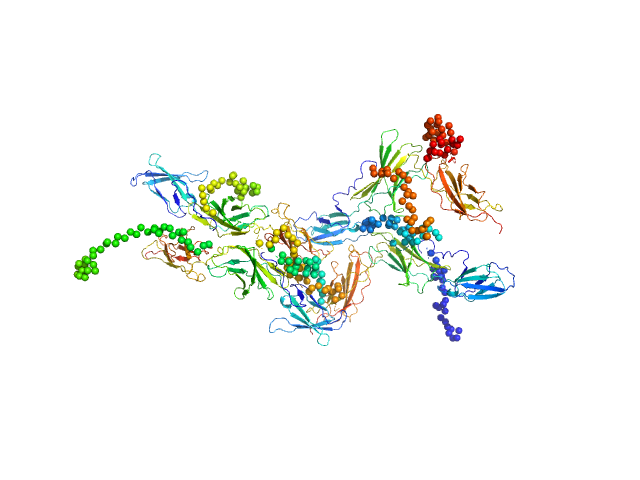
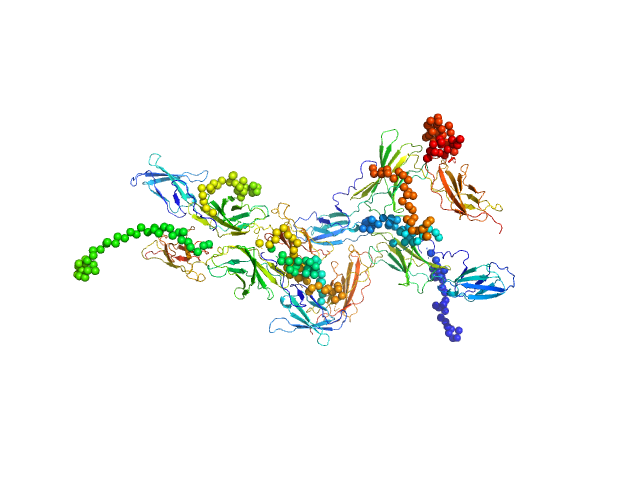
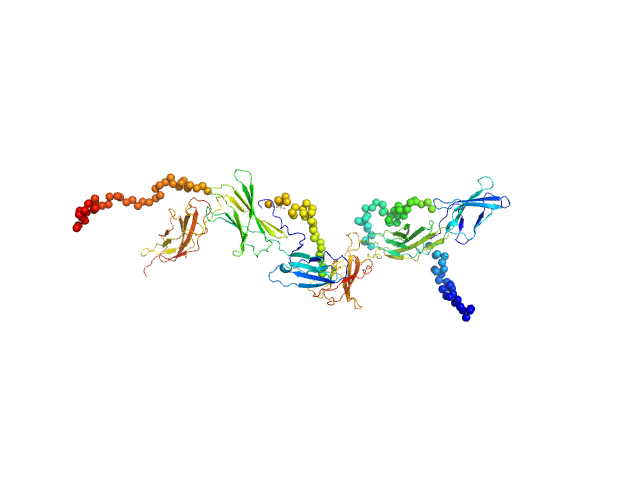
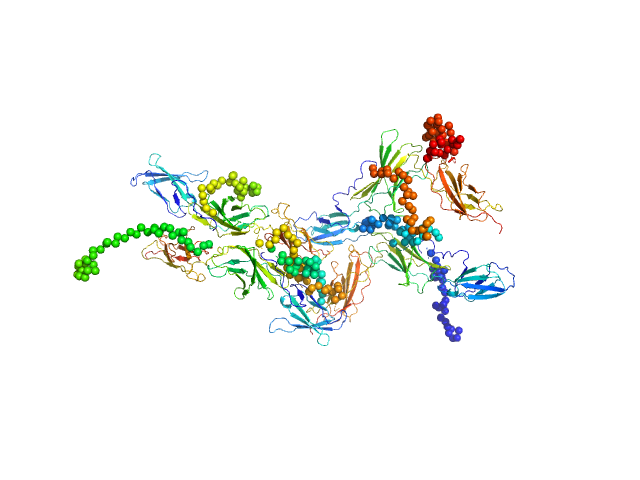
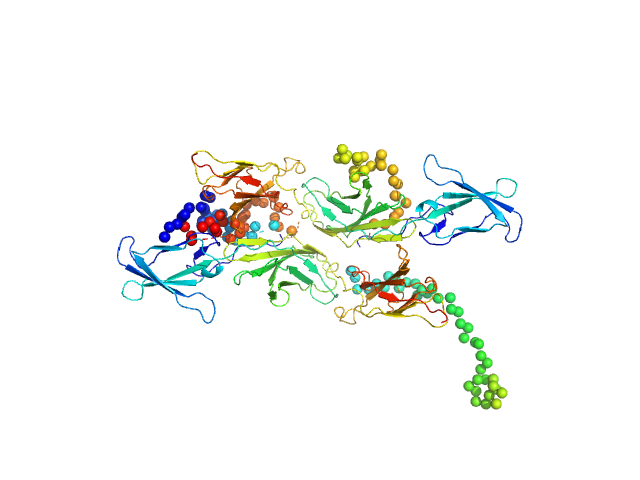
The curve was fitted with a tetramer obtained with CORAL from the unit cell tetrameric structure of the pdb code 1N26 with a Chi-squared-value of 2.49. Since the experimental MW appeared smaller than a tetramer (confirmed by a not-fitting tetrameric model) we tried to fit a mixture of tetramer-dimer using OLIGOMER. Two possible dimers were possible: a compact and an extended one. With the compact dimer mixture OLIGOMER estimated a volume fraction of 65% tetramer and 35% dimer and a corresponding Chi-squared-value of 1.92. With the extended dimer mixture OLIGOMER estimated a volume fraction of 47% tetramer and 53% dimer and a improved Chi-squared-value of 1.33. |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||