|
Synchrotron SAXS data from solutions of full length tetrameric FKBP39 protein from Drosophila melanogaster in 50 mM Na2HPO4, 150 mM NaCl, pH 7 were collected on the B21 beam line at the Diamond Light Source (Oxfordshire, UK), using a Pilatus 2M detector at a sample-detector distance of 3.9 m and at a wavelength of λ = 0.1 nm (I(s) vs s = 4π sin θ/λ, where 2θ is the scattering angle). One solute concentration of 3.00 mg/ml was measured at 10°C. 18 successive 10 second frames were collected. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
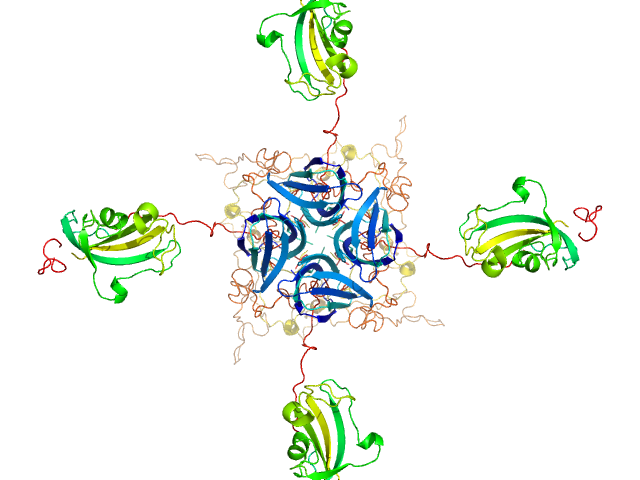
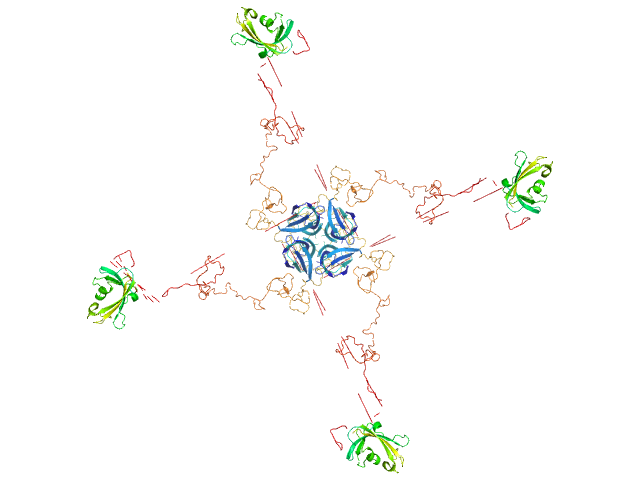
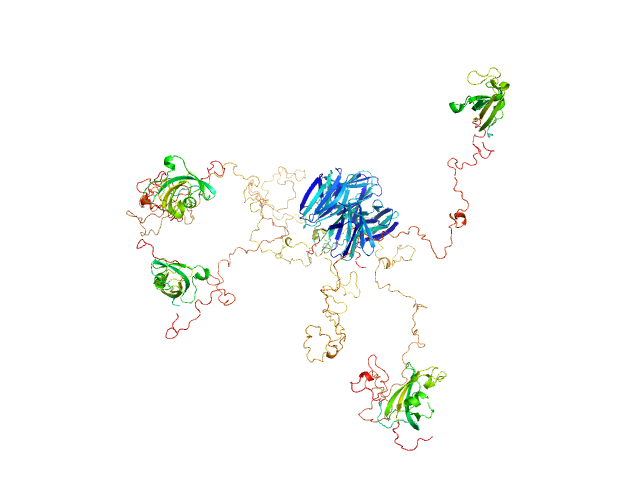
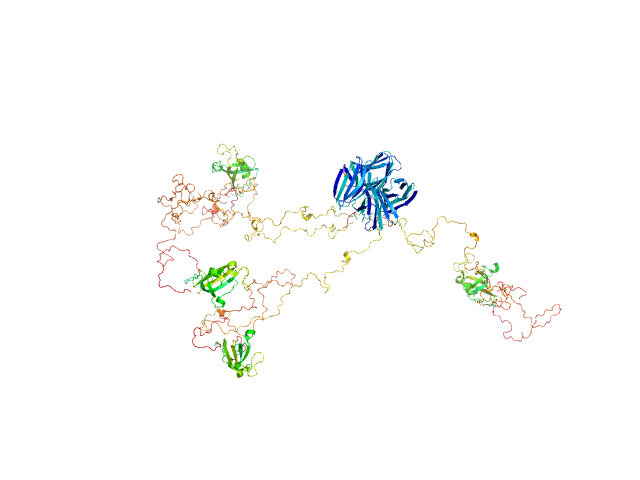
Modeling of the full length FKBP39 protein from low resolution structure was performed using Ensemble Optimization Method (EOM). EOM models were then processed by PD2 ca2main program to generate models that have replaced bead residues by C-alpha backbone. The molecular weight of the protein was estimated using I(0) value obtained for the CS domain of barley SGT1. Molecular weight obtained using MoW (Fischer et al) is 158 kDa. In addition, a zip file containing models of the final EOM ensemble as well as the final Rg and Dmax distributions are available in the full entry zip archive.
|
|
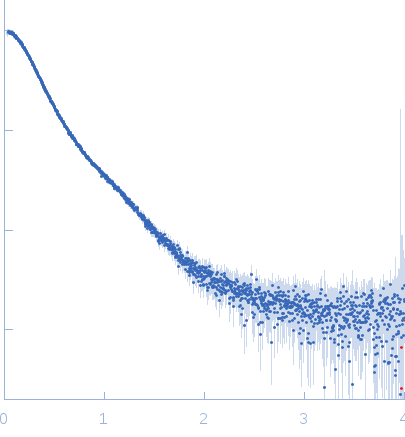 s, nm-1
s, nm-1
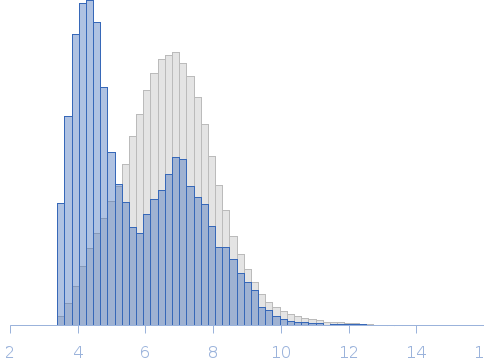 Rg, nm
Rg, nm